1FP4
 
 | | CRYSTAL STRUCTURE OF THE ALPHA-H195Q MUTANT OF NITROGENASE | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE-MO-S CLUSTER, ... | | Authors: | Sorlie, M, Christiansen, J, Lemon, B.J, Peters, J.W, Dean, D.R, Hales, B.J. | | Deposit date: | 2000-08-30 | | Release date: | 2002-11-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic features and structure of the nitrogenase alpha-Gln195 MoFe protein
Biochemistry, 40, 2001
|
|
1FP6
 
 | | THE NITROGENASE FE PROTEIN FROM AZOTOBACTER VINELANDII COMPLEXED WITH MGADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Jang, S.B, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2000-08-30 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insights into nucleotide signal transduction in nitrogenase: structure of an iron protein with MgADP bound.
Biochemistry, 39, 2000
|
|
1G20
 
 | | MGATP-BOUND AND NUCLEOTIDE-FREE STRUCTURES OF A NITROGENASE PROTEIN COMPLEX BETWEEN LEU127DEL-FE PROTEIN AND THE MOFE PROTEIN | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Chiu, H.-J, Peters, J.W, Lanzilotta, W.N, Ryle, M.J, Seefeldt, L.C, Howard, J.B, Rees, D.C. | | Deposit date: | 2000-10-16 | | Release date: | 2001-01-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MgATP-Bound and nucleotide-free structures of a nitrogenase protein complex between the Leu 127 Delta-Fe-protein and the MoFe-protein.
Biochemistry, 40, 2001
|
|
1G21
 
 | | MGATP-BOUND AND NUCLEOTIDE-FREE STRUCTURES OF A NITROGENASE PROTEIN COMPLEX BETWEEN LEU127DEL-FE PROTEIN AND THE MOFE PROTEIN | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Chiu, H.-J, Peters, J.W, Lanzilotta, W.N, Ryle, M.J, Seefeldt, L.C, Howard, J.B, Rees, D.C. | | Deposit date: | 2000-10-16 | | Release date: | 2001-01-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | MgATP-Bound and nucleotide-free structures of a nitrogenase protein complex between the Leu 127 Delta-Fe-protein and the MoFe-protein.
Biochemistry, 40, 2001
|
|
7MGN
 
 | | Crystal structure of F501H/H506E variant of 2-ketopropyl coenzyme M oxidoreductase/carboxylase (2-KPCC) from Xanthobacter autotrophicus | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-oxopropyl-CoM reductase, carboxylating, ... | | Authors: | Zadvornyy, O.A, Prussia, G, Peters, J.W. | | Deposit date: | 2021-04-12 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The unique Phe-His dyad of 2-ketopropyl coenzyme M oxidoreductase/carboxylase selectively promotes carboxylation and S-C bond cleavage.
J.Biol.Chem., 297, 2021
|
|
7MGO
 
 | | Crystal structure of F501H variant of 2-ketopropyl coenzyme M oxidoreductase/carboxylase (2-KPCC) from Xanthobacter autotrophicus | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-oxopropyl-CoM reductase, carboxylating, ... | | Authors: | Prussia, G, Zadvornyy, O.A, Peters, J.W. | | Deposit date: | 2021-04-13 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The unique Phe-His dyad of 2-ketopropyl coenzyme M oxidoreductase/carboxylase selectively promotes carboxylation and S-C bond cleavage.
J.Biol.Chem., 297, 2021
|
|
3FWY
 
 | | Crystal structure of the L protein of Rhodobacter sphaeroides light-independent protochlorophyllide reductase (BchL) with MgADP bound: a homologue of the nitrogenase Fe protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase iron-sulfur ATP-binding protein, ... | | Authors: | Sarma, R, Barney, B.M, Hamilton, T.L, Jones, A, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2009-01-19 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of the L Protein of Rhodobacter sphaeroides Light-Independent Protochlorophyllide Reductase with MgADP Bound: A Homologue of the Nitrogenase Fe Protein.
Biochemistry, 47, 2008
|
|
1RW4
 
 | | Nitrogenase Fe protein l127 deletion variant | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, Nitrogenase iron protein 1 | | Authors: | Sen, S, Igarashi, R, Smith, A, Johnson, M.K, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2003-12-15 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Conformational Mimic of the MgATP-Bound "On State" of the Nitrogenase Iron Protein.
Biochemistry, 43, 2004
|
|
3PND
 
 | | FAD binding by ApbE protein from Salmonella enterica: a new class of FAD binding proteins | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, Thiamine biosynthesis lipoprotein ApbE | | Authors: | Boyd, J.M, Endrizzi, J.A, Hamilton, T.L, Christopherson, M.R, Mulder, D.W, Downs, D.M, Peters, J.W. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | FAD binding by ApbE protein from Salmonella enterica: a new class of FAD-binding proteins.
J.Bacteriol., 193, 2011
|
|
1WKM
 
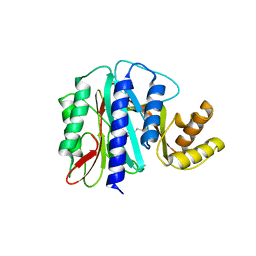 | | THE PRODUCT BOUND FORM OF THE MN(II)LOADED METHIONINE AMINOPEPTIDASE FROM HYPERTHERMOPHILE PYROCOCCUS FURIOSUS | | Descriptor: | MANGANESE (II) ION, METHIONINE, Methionine aminopeptidase | | Authors: | Copik, A.J, Nocek, B.P, Jang, S.B, Swierczek, S.I, Ruebush, S, Meng, L, D'souza, V.M, Peters, J.W, Bennett, B, Holz, R.C. | | Deposit date: | 2004-06-01 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | EPR and X-ray crystallographic characterization of the product-bound form of the MnII-loaded methionyl aminopeptidase from Pyrococcus furiosus
Biochemistry, 44, 2005
|
|
1XD8
 
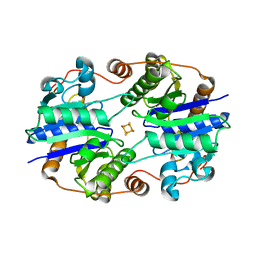 | | Crystal Structure of the Nitrogenase Fe protein Asp39Asn | | Descriptor: | IRON/SULFUR CLUSTER, Nitrogenase iron protein 1 | | Authors: | Jang, S.B, Jeong, M.S, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2004-09-05 | | Release date: | 2005-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biochemical implications of single amino acid substitutions in the nucleotide-dependent switch regions of the nitrogenase Fe protein from Azotobacter vinelandii
J.Biol.Inorg.Chem., 9, 2004
|
|
1XDB
 
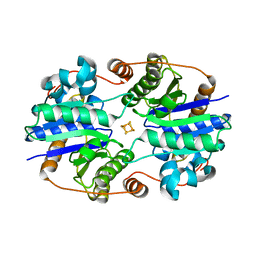 | | Crystal Structure of the Nitrogenase Fe protein Asp129Glu | | Descriptor: | IRON/SULFUR CLUSTER, Nitrogenase iron protein 1 | | Authors: | Jang, S.B, Jeong, M.S, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2004-09-05 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical implications of single amino acid substitutions in the nucleotide-dependent switch regions of the nitrogenase Fe protein from Azotobacter vinelandii
J.Biol.Inorg.Chem., 9, 2004
|
|
1XD9
 
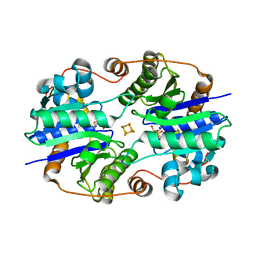 | | Crystal Structure of the Nitrogenase Fe protein Asp39Asn with MgADP bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Jang, S.B, Jeong, M.S, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2004-09-05 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical implications of single amino acid substitutions in the nucleotide-dependent switch regions of the nitrogenase Fe protein from Azotobacter vinelandii
J.Biol.Inorg.Chem., 9, 2004
|
|
1C4A
 
 | |
1C4C
 
 | |
1DE0
 
 | |
1MOK
 
 | | NADPH DEPENDENT 2-KETOPROPYL COENZYME M OXIDOREDUCTASE/CARBOXYLASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, orf3 | | Authors: | Nocek, B, Jang, S.B, Jeong, M.S, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2002-09-09 | | Release date: | 2002-11-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for CO2 Fixation by a Novel Member of the Disulfide Oxidoreductase Family of Enzymes,
2-Ketopropyl Coenzyme M Oxidoreductase/Carboxylase
Biochemistry, 41, 2002
|
|
2C3C
 
 | | 2.01 Angstrom X-ray crystal structure of a mixed disulfide between coenzyme M and NADPH-dependent oxidoreductase 2-ketopropyl coenzyme M carboxylase | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-OXOPROPYL-COM REDUCTASE, ACETONE, ... | | Authors: | Pandey, A.S, Nocek, B, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2005-10-05 | | Release date: | 2005-12-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanistic Implications of the Structure of the Mixed-Disulfide Intermediate of the Disulfide Oxidoreductase, 2-Ketopropyl-Coenzyme M Oxidoreductase/Carboxylase.
Biochemistry, 45, 2006
|
|
2C8V
 
 | | Insights into the role of nucleotide-dependent conformational change in nitrogenase catalysis: Structural characterization of the nitrogenase Fe protein Leu127 deletion variant with bound MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION, ... | | Authors: | Sen, S, Krishnakumar, A, McClead, J, Johnson, M.K, Seefeldt, L.C, Szilagyi, R.K, Peters, J.W. | | Deposit date: | 2005-12-08 | | Release date: | 2006-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights Into the Role of Nucleotide-Dependent Conformational Change in Nitrogenase Catalysis: Structural Characterization of the Nitrogenase Fe Protein Leu127 Deletion Variant with Bound Mgatp.
J.Inorg.Biochem., 100, 2006
|
|
2CFC
 
 | | structural basis for stereo selectivity in the (R)- and (S)- hydroxypropylethane thiosulfonate dehydrogenases | | Descriptor: | (2-[2-KETOPROPYLTHIO]ETHANESULFONATE, 2-(R)-HYDROXYPROPYL-COM DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Krishnakumar, A.M, Nocek, B.P, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2006-02-19 | | Release date: | 2006-07-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Stereoselectivity in the (R)-and (S)-Hydroxypropylthioethanesulfonate Dehydrogenases.
Biochemistry, 45, 2006
|
|
2C3D
 
 | | 2.15 Angstrom crystal structure of 2-ketopropyl coenzyme M oxidoreductase carboxylase with a coenzyme M disulfide bound at the active site | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-OXOPROPYL-COM REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pandey, A.S, Nocek, B, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2005-10-05 | | Release date: | 2005-11-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanistic Implications of the Structure of the Mixed-Disulfide Intermediate of the Disulfide Oxidoreductase, 2-Ketopropyl-Coenzyme M Oxidoreductase/Carboxylase.
Biochemistry, 45, 2006
|
|
5SVC
 
 | | Mechanism of ATP-Dependent Acetone Carboxylation, Acetone Carboxylase nucleotide-free structure | | Descriptor: | Acetone carboxylase alpha subunit, Acetone carboxylase beta subunit, Acetone carboxylase gamma subunit, ... | | Authors: | Eilers, B.J, Mus, F, Alleman, A.B, Kabasakal, B.V, Murray, J.W, Nocek, B.P, Dubois, J.L, Peters, J.W. | | Deposit date: | 2016-08-05 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Mechanism of ATP-Dependent Acetone Carboxylation.
Sci Rep, 7, 2017
|
|
5SVB
 
 | | Mechanism of ATP-Dependent Acetone Carboxylation, Acetone Carboxylase AMP bound structure | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acetone carboxylase alpha subunit, Acetone carboxylase beta subunit, ... | | Authors: | Eilers, B.J, Mus, F, Alleman, A.B, Kabasakal, B.V, Murray, J.W, Nocek, B.P, Dubois, J.L, Peters, J.W. | | Deposit date: | 2016-08-05 | | Release date: | 2017-08-09 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Structural Basis for the Mechanism of ATP-Dependent Acetone Carboxylation.
Sci Rep, 7, 2017
|
|
5T2K
 
 | | Geobacillus stearothermophilus HemQ with Manganese-Coproporphyrin III | | Descriptor: | Putative heme-dependent peroxidase GT50_08830, [3,3',3'',3'''-(3,8,13,17-tetramethylporphyrin-2,7,12,18-tetrayl-kappa~4~N~21~,N~22~,N~23~,N~24~)tetra(propanoato)(2-)]manganese | | Authors: | Gauss, G.H, Celis, A.I, Dubois, J.L, Peters, J.W. | | Deposit date: | 2016-08-23 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Mechanism for Oxidative Decarboxylation Reactions Mediated by Amino Acids and Heme Propionates in Coproheme Decarboxylase (HemQ).
J. Am. Chem. Soc., 139, 2017
|
|
5VJ7
 
 | | Ferredoxin NADP Oxidoreductase (Xfn) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin-NADP(+) reductase subunit alpha, ... | | Authors: | Zadvornyy, O.A, Nguyen, D.M.N, Schut, G.J, Lipscomb, G.L, Tokmina-Lukaszewska, M, Adams, M.W.W, Peters, J.W. | | Deposit date: | 2017-04-18 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Two functionally distinct NADP(+)-dependent ferredoxin oxidoreductases maintain the primary redox balance of Pyrococcus furiosus.
J. Biol. Chem., 292, 2017
|
|
