8GDC
 
 | | Cryo-EM Structure of the Prostaglandin E2 Receptor 3 Coupled to G Protein | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Shenming, H, Mengyao, X, Lei, L, Yang, D, Shiyi, G, Jinpeng, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GD9
 
 | | Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein | | Descriptor: | 4-({2-[(1R,2R,3R,5S)-3,5-dihydroxy-2-{(3S)-3-hydroxy-4-[3-(methoxymethyl)phenyl]butyl}cyclopentyl]ethyl}sulfanyl)butanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Shenming, H, Mengyao, X, Lei, L, Yang, D, Sheng, C, Jinpeng, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3AHT
 
 | | Crystal structure of rice BGlu1 E176Q mutant in complex with laminaribiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Chuenchor, W, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Svasti, J, Ketudat Cairns, J.R. | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3AHV
 
 | | Semi-active E176Q mutant of rice bglu1 covalent complex with 2-deoxy-2-fluoroglucoside | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-alpha-D-glucopyranose, Beta-glucosidase 7, ... | | Authors: | Chuenchor, W, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Svasti, J, Ketudat Cairns, J.R. | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3F5K
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3F5J
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, SULFATE ION, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3F5L
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, SULFATE ION, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-04 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3F4V
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
7W2W
 
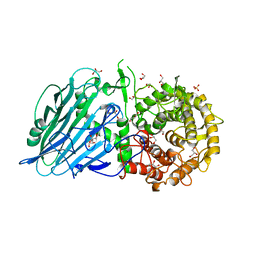 | | Crystal structure of TxGH116 R786K mutant from Thermoanaerobacterium xylanolyticum with glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
6K98
 
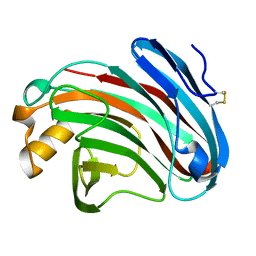 | | Substrates promiscuity of xyloglucanases and endoglucanases of glycoside hydrolase 12 family | | Descriptor: | GH12 beta-1, 4-endoglucanase | | Authors: | Hong, Y, Tao, T, Pengjun, S, Jiaming, C, Xiaoyu, W, Chen, H, yingguo, B, Bin, Y. | | Deposit date: | 2019-06-14 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | Substrates promiscuity of xyloglucanases and endoglucanases of glycoside hydrolase 12 family
To Be Published
|
|
6K9D
 
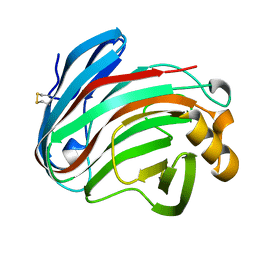 | | glycoside hydrolase family 12 (GH12) englucanase | | Descriptor: | GH12 beta-1, 4-endoglucanase | | Authors: | Hong, Y, Tao, T, Pengjun, S, Jiaming, C, Xiaoyu, W, Chen, H, Yingguo, B, Bin, Y. | | Deposit date: | 2019-06-14 | | Release date: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.505 Å) | | Cite: | Substrates promiscuity of xyloglucanases and endoglucanases of glycoside hydrolase 12 family
To Be Published
|
|
7W2X
 
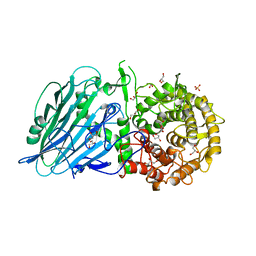 | | Crystal structure of TxGH116 R786K mutant from Thermoanaerobacterium xylanolyticum | | Descriptor: | CALCIUM ION, GLYCEROL, Glucosylceramidase, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
7W2S
 
 | | Crystal structure of TxGH116 E730A mutant from Thermoanaerobacterium xylanolyticum with glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
7W2T
 
 | | Crystal structure of TxGH116 E730Q mutant from Thermoanaerobacterium xylanolyticum with glucose | | Descriptor: | CALCIUM ION, GLYCEROL, Glucosylceramidase, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
7W2V
 
 | | Crystal structure of TxGH116 R786A mutant from Thermoanaerobacterium xylanolyticum with glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
