7MXZ
 
 | | Sy-CrtE apo structure | | Descriptor: | CHLORIDE ION, Geranylgeranyl pyrophosphate synthase, MAGNESIUM ION | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2021-05-19 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Molecular characterization of cyanobacterial short-chain prenyltransferases and discovery of a novel GGPP phosphatase.
Febs J., 289, 2022
|
|
7MY7
 
 | | Se-CrtE N-term His-tag structure | | Descriptor: | Farnesyl-diphosphate synthase | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2021-05-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Molecular characterization of cyanobacterial short-chain prenyltransferases and discovery of a novel GGPP phosphatase.
Febs J., 289, 2022
|
|
6C7W
 
 | | Carbonic anhydrase 2 in complex with [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDRO-2-FURANYL]METHYL SULFAMATE inhibitor | | Descriptor: | Carbonic anhydrase 2, SODIUM ION, ZINC ION, ... | | Authors: | Peat, T.S, Mujumdar, P, Poulsen, S.A. | | Deposit date: | 2018-01-23 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Synthesis, structure and bioactivity of primary sulfamate-containing natural products.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6C7X
 
 | | Carbonic anhydrase 2 in complex with 2-chloro-5'-O-sulfamoyladenosine | | Descriptor: | 2-chloro-5'-O-sulfamoyladenosine, Carbonic anhydrase 2, SODIUM ION, ... | | Authors: | Peat, T.S, Mujumdar, P, Poulsen, S.A. | | Deposit date: | 2018-01-23 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis, structure and bioactivity of primary sulfamate-containing natural products.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6C62
 
 | | An unexpected vestigial protein complex reveals the evolutionary origins of an s-triazine catabolic enzyme. | | Descriptor: | AtzG, Biuret hydrolase, MAGNESIUM ION | | Authors: | Peat, T.S, Esquirol, L, Wilding, M, Liu, J.W, French, N.G, Hartley, C.J, Hideki, O, Easton, C.J, Newman, J, Scott, C. | | Deposit date: | 2018-01-17 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An unexpected vestigial protein complex reveals the evolutionary origins of ans-triazine catabolic enzyme.
J. Biol. Chem., 293, 2018
|
|
6C6G
 
 | | An unexpected vestigial protein complex reveals the evolutionary origins of an s-triazine catabolic enzyme. Inhibitor bound complex. | | Descriptor: | AtzG, Biuret hydrolase, CALCIUM ION | | Authors: | Peat, T.S, Esquirol, L, Wilding, M, Liu, J.W, French, N.G, Hartley, C.J, Hideki, O, Easton, C.J, Newman, J, Scott, C. | | Deposit date: | 2018-01-18 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An unexpected vestigial protein complex reveals the evolutionary origins of ans-triazine catabolic enzyme.
J. Biol. Chem., 293, 2018
|
|
3NF8
 
 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | Descriptor: | 6-[(5-chloro-2-oxo-2,3-dihydro-1H-indol-1-yl)methyl]-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, ACETIC ACID, Integrase, ... | | Authors: | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
6AZN
 
 | | Structural and biochemical characterization of a non-canonical biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Putative amidase | | Authors: | Peat, T.S, Esquirol, L, Newman, J, Scott, C. | | Deposit date: | 2017-09-11 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical characterization of the biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841.
PLoS ONE, 13, 2018
|
|
6AZQ
 
 | | Structural and biochemical characterization of a non-canonical biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841 | | Descriptor: | CALCIUM ION, Putative amidase, dicarbonimidic diamide | | Authors: | Peat, T.S, Esquirol, L, Newman, J, Scott, C. | | Deposit date: | 2017-09-11 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and biochemical characterization of the biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841.
PLoS ONE, 13, 2018
|
|
6BJT
 
 | | The structure of AtzH: a little known member of the atrazine breakdown pathway | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peat, T.S, Newman, J, Scott, C, Esquirol, L. | | Deposit date: | 2017-11-07 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel decarboxylating amidohydrolase involved in avoiding metabolic dead ends during cyanuric acid catabolism in Pseudomonas sp. strain ADP.
PLoS ONE, 13, 2018
|
|
6AZS
 
 | | Structural and biochemical characterization of a non-canonical biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peat, T.S, Esquirol, L, Newman, J, Scott, C. | | Deposit date: | 2017-09-11 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural and biochemical characterization of the biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841.
PLoS ONE, 13, 2018
|
|
6AZO
 
 | | Structural and biochemical characterization of a non-canonical biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841 | | Descriptor: | CHLORIDE ION, Putative amidase | | Authors: | Peat, T.S, Esquirol, L, Newman, J, Scott, C. | | Deposit date: | 2017-09-11 | | Release date: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and biochemical characterization of the biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841.
PLoS ONE, 13, 2018
|
|
6BJU
 
 | |
3AVG
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AVH
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AVL
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AV9
 
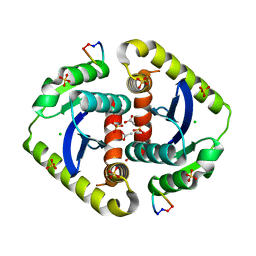 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-02 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AVF
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AVN
 
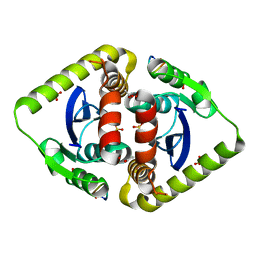 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AVI
 
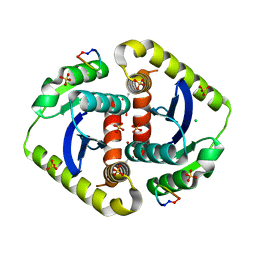 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-02-15 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AVB
 
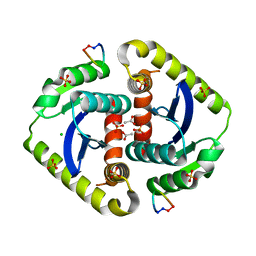 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-02 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AVJ
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AVK
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AVM
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
1UMU
 
 | |
