3P41
 
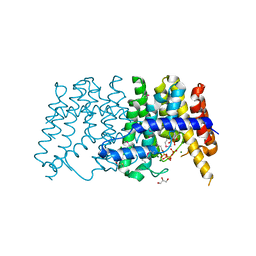 | | Crystal structure of polyprenyl synthetase from pseudomonas fluorescens pf-5 complexed with magnesium and isoprenyl pyrophosphate | | 分子名称: | CHLORIDE ION, DIMETHYLALLYL DIPHOSPHATE, GLYCEROL, ... | | 著者 | Patskovsky, Y, Toro, R, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-10-05 | | 公開日 | 2010-10-20 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3N3D
 
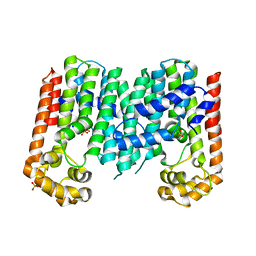 | | Crystal structure of geranylgeranyl pyrophosphate synthase from lactobacillus brevis atcc 367 | | 分子名称: | Geranylgeranyl pyrophosphate synthase, SULFATE ION | | 著者 | Patskovsky, Y, Toro, R, Rutter, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-05-19 | | 公開日 | 2010-06-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase from Lactobacillus Brevis
To be Published
|
|
2O16
 
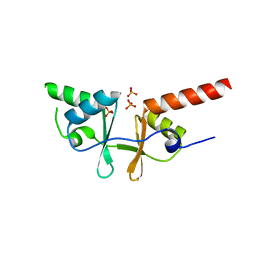 | | Crystal structure of a putative acetoin utilization protein (AcuB) from Vibrio cholerae | | 分子名称: | Acetoin utilization protein AcuB, putative, PHOSPHATE ION | | 著者 | Patskovsky, Y, Bonanno, J.B, Rutter, M, Bain, K.T, Powell, A, Slocombe, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2006-11-28 | | 公開日 | 2006-12-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of a putative acetoin utilization protein (AcuB) from Vibrio cholerae
To be Published
|
|
3PDE
 
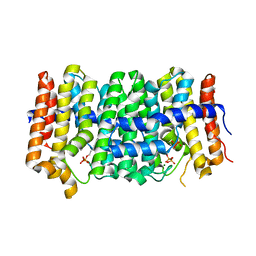 | | Crystal structure of geranylgeranyl pyrophosphate synthase from Lactobacillus brevis atcc 367 complexed with isoprenyl diphosphate and magnesium | | 分子名称: | DIMETHYLALLYL DIPHOSPHATE, Farnesyl-diphosphate synthase, GLYCEROL, ... | | 著者 | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-10-22 | | 公開日 | 2010-11-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2OOG
 
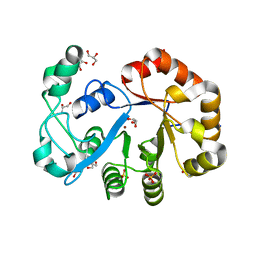 | | Crystal structure of glycerophosphoryl diester phosphodiesterase from Staphylococcus aureus | | 分子名称: | GLYCEROL, Glycerophosphoryl diester phosphodiesterase, SULFATE ION, ... | | 著者 | Patskovsky, Y, Fedorov, E, Toro, R, Sauder, J.M, Smith, D, Freeman, J, Maletic, M, Powell, A, Gheyi, T, Wasserman, S.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-01-25 | | 公開日 | 2007-02-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of Glycerophosphoryl Diester Phosphodiesterase from Staphylococcus Aureus
To be Published
|
|
2P0I
 
 | | Crystal structure of L-rhamnonate dehydratase from Gibberella zeae | | 分子名称: | GLYCEROL, L-rhamnonate dehydratase, SULFATE ION | | 著者 | Patskovsky, Y, Toro, R, Sauder, J.M, Dickey, M, Logan, C, Gheyi, T, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-02-28 | | 公開日 | 2007-03-13 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of L-Rhamnonate Dehydratase from Gibberella Zeae
To be Published
|
|
2O56
 
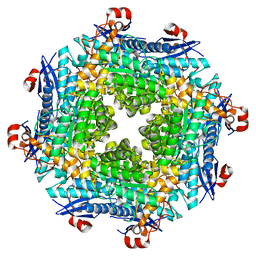 | | Crystal Structure of a Member of the Enolase Superfamily from Salmonella Typhimurium | | 分子名称: | MAGNESIUM ION, Putative mandelate racemase | | 著者 | Patskovsky, Y, Sauder, J.M, Dickey, M, Adams, J.M, Ozyurt, S, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2006-12-05 | | 公開日 | 2006-12-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Putative Enolase from Salmonella Typhimurium Lt2
To be Published
|
|
2O8R
 
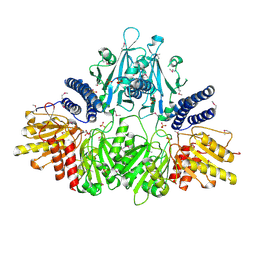 | | Crystal Structure of Polyphosphate Kinase from Porphyromonas Gingivalis | | 分子名称: | Polyphosphate kinase, SULFATE ION | | 著者 | Patskovsky, Y, Toro, R, Sauder, J.M, Dickey, M, Adams, J.M, Ozyurt, S, Wasserman, S.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2006-12-12 | | 公開日 | 2006-12-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal Structure of Polyphosphate Kinase from Porphyromonas Gingivalis
To be Published
|
|
3NF2
 
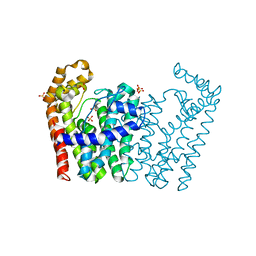 | | Crystal structure of polyprenyl synthetase from Streptomyces coelicolor A3(2) | | 分子名称: | Putative polyprenyl synthetase, SULFATE ION | | 著者 | Patskovsky, Y, Toro, R, Dickey, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-06-09 | | 公開日 | 2010-08-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3NFU
 
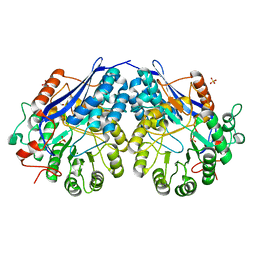 | | Crystal structure of probable glucarate dehydratase from chromohalobacter salexigens dsm 3043 complexed with magnesium | | 分子名称: | GLYCEROL, Glucarate dehydratase, MAGNESIUM ION, ... | | 著者 | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Gerlt, J.A, Almo, S.C, Burley, S.K, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-06-10 | | 公開日 | 2010-06-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Crystal Structure of Glucarate Dehydratase from Chromohalobacter Salexigens
To be Published
|
|
3N2C
 
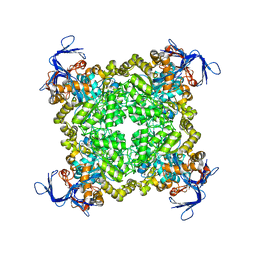 | | Crystal structure of prolidase eah89906 complexed with n-methylphosphonate-l-proline | | 分子名称: | 1-[(R)-hydroxy(methyl)phosphoryl]-L-proline, PROLIDASE, ZINC ION | | 著者 | Patskovsky, Y, Xu, C, Sauder, J.M, Burley, S.K, Raushel, F.M, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-05-17 | | 公開日 | 2010-06-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
2POD
 
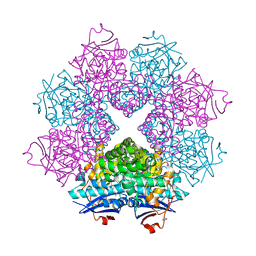 | | Crystal structure of a member of enolase superfamily from Burkholderia pseudomallei K96243 | | 分子名称: | Mandelate racemase / muconate lactonizing enzyme, SODIUM ION | | 著者 | Patskovsky, Y, Bonanno, J, Sauder, J.M, Gilmore, J.M, Iizuka, M, Ozyurt, S, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-04-26 | | 公開日 | 2007-06-05 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Crystal Structure of a Member of Enolase Superfamily from Burkholderia Pseudomallei.
To be Published
|
|
3Q2Q
 
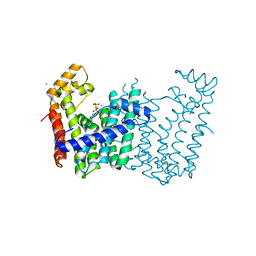 | | Crystal structure of geranylgeranyl pyrophosphate synthase from Corynebacterium glutamicum complexed with calcium and isoprenyl diphosphate | | 分子名称: | CALCIUM ION, DIMETHYLALLYL DIPHOSPHATE, GLYCEROL, ... | | 著者 | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Poulter, C.D, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-12-20 | | 公開日 | 2011-01-12 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2PMB
 
 | | Crystal structure of predicted nucleotide-binding protein from Vibrio cholerae | | 分子名称: | GLYCEROL, PHOSPHATE ION, Uncharacterized protein | | 著者 | Patskovsky, Y, Zhan, C, Shi, W, Toro, R, Sauder, J.M, Gilmore, J, Iizuka, M, Maletic, M, Gheyi, T, Wasserman, S.R, Smith, D, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-04-20 | | 公開日 | 2007-05-08 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Crystal Structure of Predicted Nucleotide-Binding Protein from Vibrio Cholerae.
To be Published
|
|
2OX4
 
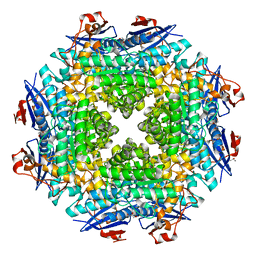 | | Crystal structure of putative dehydratase from Zymomonas mobilis ZM4 | | 分子名称: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Patskovsky, Y, Toro, R, Sauder, J.M, Freeman, J.C, Bain, K, Gheyi, T, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-02-19 | | 公開日 | 2007-03-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of Putative Dehydratase from Zymomonas Mobilis Zm4
To be Published
|
|
2OZ3
 
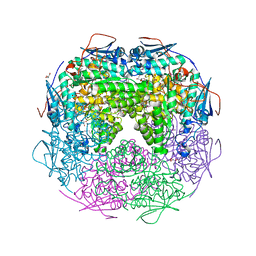 | | Crystal structure of L-Rhamnonate dehydratase from Azotobacter vinelandii | | 分子名称: | GLYCEROL, Mandelate racemase/muconate lactonizing enzyme, SODIUM ION | | 著者 | Patskovsky, Y, Toro, R, Sauder, J.M, Freeman, J.C, Bain, K, Gheyi, T, Wu, B, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-02-23 | | 公開日 | 2007-03-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of L-Rhamnonate dehydratase from azotobacter vinelandii
To be Published
|
|
3P8R
 
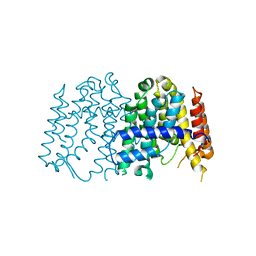 | | CRYSTAL STRUCTURE OF POLYPRENYL SYNTHASE FROM Vibrio cholerae | | 分子名称: | Geranyltranstransferase | | 著者 | Patskovsky, Y, Toro, R, Dickey, M, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-10-14 | | 公開日 | 2010-10-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3PKO
 
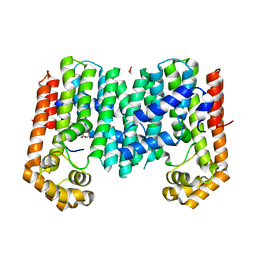 | | Crystal structure of geranylgeranyl pyrophosphate synthase from lactobacillus brevis atcc 367 complexed with citrate | | 分子名称: | CITRIC ACID, GLYCEROL, Geranylgeranyl pyrophosphate synthase | | 著者 | Patskovsky, Y, Toro, R, Rutter, M, Chang, S, Sauder, J.M, Poulter, C.D, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-11-11 | | 公開日 | 2010-11-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3Q1O
 
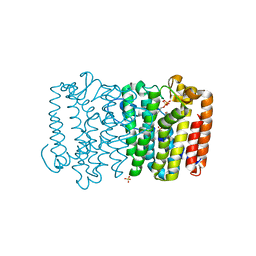 | | Crystal structure of geranyltransferase from helicobacter pylori complexed with magnesium and isoprenyl diphosphate | | 分子名称: | DIMETHYLALLYL DIPHOSPHATE, Geranyltranstransferase (IspA), MAGNESIUM ION, ... | | 著者 | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-12-17 | | 公開日 | 2011-01-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3MWC
 
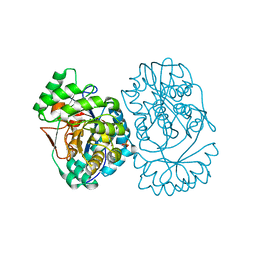 | | Crystal structure of probable o-succinylbenzoic acid synthetase from kosmotoga olearia | | 分子名称: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein, PHOSPHATE ION | | 著者 | Patskovsky, Y, Toro, R, Dickey, M, Sauder, J.M, Gerlt, J, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-05-05 | | 公開日 | 2010-05-19 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of O-Succinylbenzoic Acid Synthetase from Kosmotoga Olearia
To be Published
|
|
3MZN
 
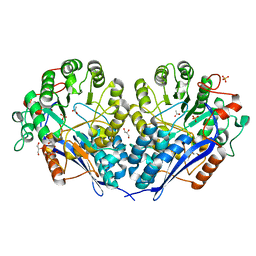 | | Crystal structure of probable glucarate dehydratase from chromohalobacter salexigens dsm 3043 | | 分子名称: | ACETATE ION, GLYCEROL, Glucarate dehydratase, ... | | 著者 | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Gerlt, J.A, Almo, S.C, Burley, S.K, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-05-12 | | 公開日 | 2010-05-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal Structure of Glucarate Dehydratase from Chromohalobacter Salexigens
To be Published
|
|
3N28
 
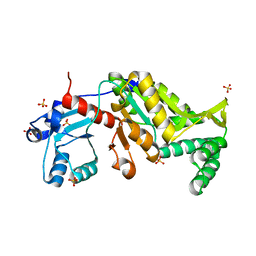 | | Crystal structure of probable phosphoserine phosphatase from vibrio cholerae, unliganded form | | 分子名称: | Phosphoserine phosphatase, SULFATE ION | | 著者 | Patskovsky, Y, Ramagopal, U, Toro, R, Rutter, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-05-17 | | 公開日 | 2010-07-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure of Phosphoserine Phosphatase from Vibrio Cholerae
To be Published
|
|
2Q01
 
 | | Crystal structure of glucuronate isomerase from Caulobacter crescentus | | 分子名称: | POTASSIUM ION, Uronate isomerase | | 著者 | Patskovsky, Y, Bonanno, J, Sridhar, V, Sauder, J.M, Freeman, J, Powell, A, Koss, J, Groshong, C, Gheyi, T, Wasserman, S.R, Raushel, F, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-05-18 | | 公開日 | 2007-05-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Crystal Structure of Glucuronate Isomerase from Caulobacter crescentus.
To be Published
|
|
2OLA
 
 | | Crystal structure of O-succinylbenzoic acid synthetase from Staphylococcus aureus, cubic crystal form | | 分子名称: | O-succinylbenzoic acid synthetase | | 著者 | Patskovsky, Y, Sauder, J.M, Ozyurt, S, Wasserman, S.R, Smith, D, Dickey, M, Maletic, M, Reyes, C, Gheyi, T, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-01-18 | | 公開日 | 2007-02-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3NRJ
 
 | | Crystal structure of probable yrbi family phosphatase from pseudomonas syringae pv.phaseolica 1448a complexed with magnesium | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Patskovsky, Y, Ramagopal, U, Toro, R, Freeman, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-06-30 | | 公開日 | 2010-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of Had Family Hydrolase from Pseudomonas Syringae Pv.Phaseolica 1448A
To be Published
|
|
