3BZM
 
 | | Crystal Structure of Open form of Menaquinone-Specific Isochorismate Synthase, MenF | | Descriptor: | CITRIC ACID, Menaquinone-specific isochorismate synthase | | Authors: | Parsons, J.F, Shi, K.M, Ladner, J.E. | | Deposit date: | 2008-01-18 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of isochorismate synthase in complex with magnesium.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
5FWG
 
 | | TETRA-(5-FLUOROTRYPTOPHANYL)-GLUTATHIONE TRANSFERASE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, TETRA-(5-FLUOROTRYPTOPHANYL)-GLUTATHIONE TRANSFERASE MU CLASS | | Authors: | Parsons, J.F, Xiao, G, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1997-11-08 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymes harboring unnatural amino acids: mechanistic and structural analysis of the enhanced catalytic activity of a glutathione transferase containing 5-fluorotryptophan.
Biochemistry, 37, 1998
|
|
1T6K
 
 | | Crystal structure of phzF from Pseudomonas fluorescens 2-79 | | Descriptor: | Phenazine biosynthesis protein phzF, SULFATE ION | | Authors: | Parsons, J.F, Song, F, Parsons, L, Calabrese, K, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2004-05-06 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of the phenazine biosynthesis protein PhzF from Pseudomonas fluorescens 2-79
Biochemistry, 43, 2004
|
|
1K0E
 
 | | THE CRYSTAL STRUCTURE OF AMINODEOXYCHORISMATE SYNTHASE FROM FORMATE GROWN CRYSTALS | | Descriptor: | FORMIC ACID, TRYPTOPHAN, p-aminobenzoate synthase component I | | Authors: | Parsons, J.F, Jensen, P.Y, Pachikara, A.S, Howard, A.J, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Escherichia coli aminodeoxychorismate synthase: architectural conservation and diversity in chorismate-utilizing enzymes.
Biochemistry, 41, 2002
|
|
1K0G
 
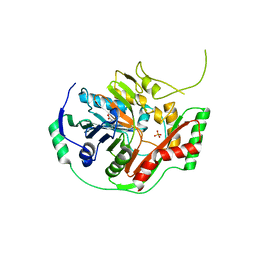 | | THE CRYSTAL STRUCTURE OF AMINODEOXYCHORISMATE SYNTHASE FROM PHOSPHATE GROWN CRYSTALS | | Descriptor: | PHOSPHATE ION, TRYPTOPHAN, p-aminobenzoate synthase component I | | Authors: | Parsons, J.F, Jensen, P.Y, Pachikara, A.S, Howard, A.J, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Escherichia coli aminodeoxychorismate synthase: architectural conservation and diversity in chorismate-utilizing enzymes.
Biochemistry, 41, 2002
|
|
1T9M
 
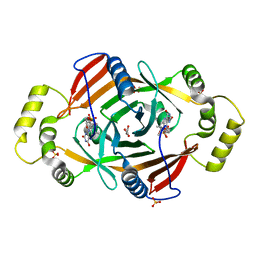 | | X-ray crystal structure of phzG from pseudomonas aeruginosa | | Descriptor: | ACETIC ACID, FLAVIN MONONUCLEOTIDE, SULFATE ION, ... | | Authors: | Parsons, J.F, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2004-05-18 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the phenazine biosynthesis enzyme PhzG.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1TY9
 
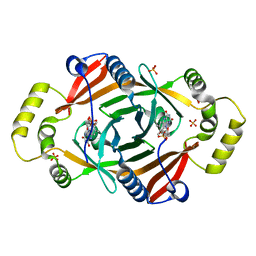 | | X-RAY CRYSTAL STRUCTURE OF PHZG FROM PSEUDOMONAS FLUORESCENS | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phenazine biosynthesis protein phzG, SULFATE ION | | Authors: | Parsons, J.F, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2004-07-07 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the phenazine biosynthesis enzyme PhzG.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3BZN
 
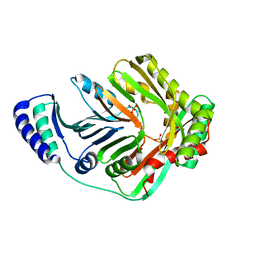 | | Crystal Structure of Open form of Menaquinone-Specific Isochorismate Synthase, MenF | | Descriptor: | MAGNESIUM ION, Menaquinone-specific isochorismate synthase, SULFATE ION | | Authors: | Parsons, J.F, Shi, K.M, Ladner, J.E. | | Deposit date: | 2008-01-18 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of isochorismate synthase in complex with magnesium.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1XAX
 
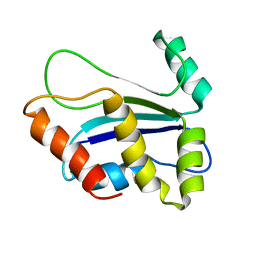 | | NMR structure of HI0004, a putative essential gene product from Haemophilus influenzae | | Descriptor: | Hypothetical UPF0054 protein HI0004 | | Authors: | Yeh, D.C, Parsons, J.F, Parsons, L.M, Liu, F, Eisenstein, E, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2004-08-26 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of HI0004, a putative essential gene product from Haemophilus influenzae, and comparison with the X-ray structure of an Aquifex aeolicus homolog
Protein Sci., 14, 2005
|
|
4GRH
 
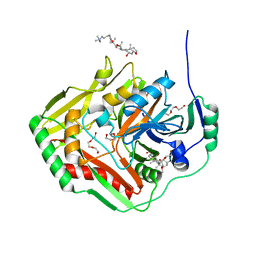 | | Crystal structure of pabB of Stenotrophomonas maltophilia | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, MAGNESIUM ION, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Bera, A, Atanasova, V, Ladner, J.E, Parsons, J.F. | | Deposit date: | 2012-08-24 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Aminodeoxychorismate Synthase from Stenotrophomonas maltophilia.
Biochemistry, 51, 2012
|
|
4HPP
 
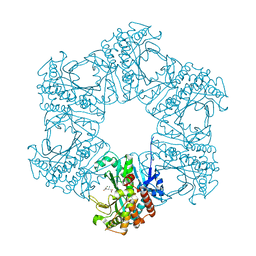 | | Crystal structure of novel glutamine synthase homolog | | Descriptor: | CALCIUM ION, GLUTAMIC ACID, MAGNESIUM ION, ... | | Authors: | Ladner, J.E, Atanasova, V, Dolezelova, Z, Parsons, J.F. | | Deposit date: | 2012-10-24 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Activity of PA5508, a Hexameric Glutamine Synthetase Homologue.
Biochemistry, 51, 2012
|
|
3L2K
 
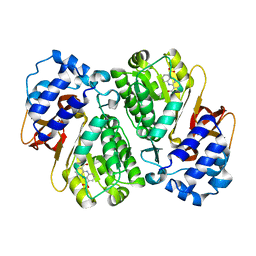 | | Structure of phenazine antibiotic biosynthesis protein with substrate | | Descriptor: | EhpF, phenazine-1,6-dicarboxylic acid | | Authors: | Bera, A.K, Atanasova, V, Parsons, J.F. | | Deposit date: | 2009-12-15 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the D-alanylgriseoluteic acid biosynthetic protein EhpF, an atypical member of the ANL superfamily of adenylating enzymes.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1I2K
 
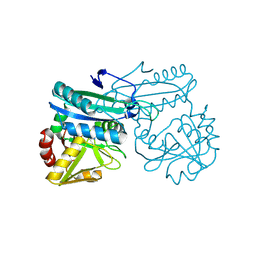 | | AMINODEOXYCHORISMATE LYASE FROM ESCHERICHIA COLI | | Descriptor: | 4-AMINO-4-DEOXYCHORISMATE LYASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jensen, P.Y, Parsons, J.F, Fisher, K.E, Pachikara, A.S, Tordova, M, Howard, A.J, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2001-02-09 | | Release date: | 2003-09-02 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure and Mechanism of Escherichia coli Aminodeoxychorismate Lyase
To be Published
|
|
1J8B
 
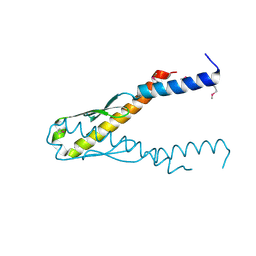 | | Structure of YbaB from Haemophilus influenzae (HI0442), a protein of unknown function | | Descriptor: | YbaB | | Authors: | Lim, K, Tempcyzk, A, Toedt, J, Parsons, J.F, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-21 | | Release date: | 2003-01-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of YbaB from Haemophilus influenzae (HI0442), a
protein of unknown function coexpressed with the recombinational
DNA repair protein RecR
Proteins, 50, 2003
|
|
1I2L
 
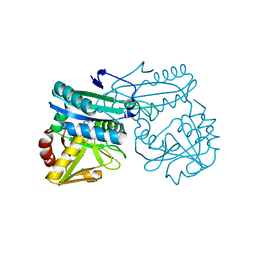 | | DEOXYCHORISMATE LYASE FROM ESCHERICHIA COLI WITH INHIBITOR | | Descriptor: | 4-AMINO-4-DEOXYCHORISMATE LYASE, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Jensen, P.Y, Parsons, J.F, Fisher, K.E, Pachikara, A.S, Tordova, M, Howard, A.J, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2001-02-09 | | Release date: | 2003-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of Escherichia coli Aminodeoxychorismate Lyase
To be Published
|
|
1JOE
 
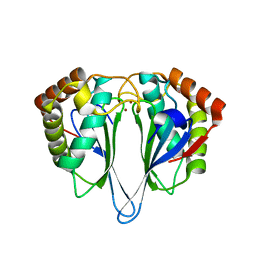 | | Crystal Structure of Autoinducer-2 Production Protein (LuxS) from Heamophilus influenzae | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN, MERCURY (II) ION, ZINC ION | | Authors: | Chen, C.C.H, Parsons, J.F, Lim, K, Lehmann, C, Tempczyk, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-27 | | Release date: | 2001-08-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CRYSTAL STRUCTURE OF AUTOINDUCER-2 PRODUCTION PROTEIN (LUXS) FROM HEAMOPHILUS INFLUENZAE--A CASE OF TWINNED CRYSTAL
To be Published
|
|
2IP2
 
 | |
3FYG
 
 | | CRYSTAL STRUCTURE OF TETRADECA-(3-FLUOROTYROSYL)-GLUTATHIONE S-TRANSFERASE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, MU CLASS TETRADECA-(3-FLUOROTYROSYL)-GLUTATHIONE S-TRANSFERASE OF ISOENZYME | | Authors: | Xiao, G, Parsons, J.F, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1997-08-07 | | Release date: | 1999-06-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational changes in the crystal structure of rat glutathione transferase M1-1 with global substitution of 3-fluorotyrosine for tyrosine.
J.Mol.Biol., 281, 1998
|
|
2RGJ
 
 | | Crystal structure of flavin-containing monooxygenase PhzS | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase | | Authors: | Ladner, J.E, Parsons, J.F, Greenhagen, B.T, Robinson, H. | | Deposit date: | 2007-10-03 | | Release date: | 2008-05-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Pyocyanin Biosynthetic Protein PhzS.
Biochemistry, 47, 2008
|
|
1N2A
 
 | | Crystal Structure of a Bacterial Glutathione Transferase from Escherichia coli with Glutathione Sulfonate in the Active Site | | Descriptor: | GLUTATHIONE SULFONIC ACID, Glutathione S-transferase | | Authors: | Rife, C.L, Parsons, J.F, Xiao, G, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2002-10-22 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conserved structural elements in glutathione transferase homologues encoded in the genome of Escherichia coli
Proteins, 53, 2003
|
|
1R4W
 
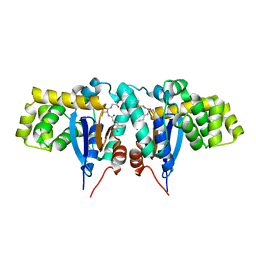 | | Crystal structure of Mitochondrial class kappa glutathione transferase | | Descriptor: | GLUTATHIONE, Glutathione S-transferase, mitochondrial | | Authors: | Ladner, J.E, Parsons, J.F, Rife, C.L, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2003-10-08 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Parallel Evolutionary Pathways for Glutathione Transferases: Structure and Mechanism of the Mitochondrial Class Kappa Enzyme rGSTK1-1
Biochemistry, 43, 2004
|
|
3H77
 
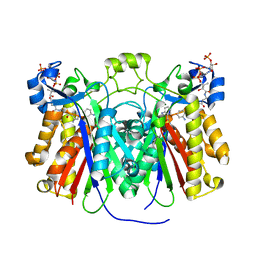 | | Crystal structure of Pseudomonas aeruginosa PqsD in a covalent complex with anthranilate | | Descriptor: | Anthraniloyl-coenzyme A, PQS biosynthetic enzyme | | Authors: | Bera, A.K, Atanasova, V, Parsons, J.F. | | Deposit date: | 2009-04-24 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of PqsD, a Pseudomonas quinolone signal biosynthetic enzyme, in complex with anthranilate.
Biochemistry, 48, 2009
|
|
3HGU
 
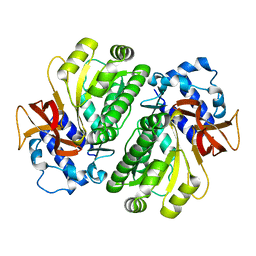 | |
3H78
 
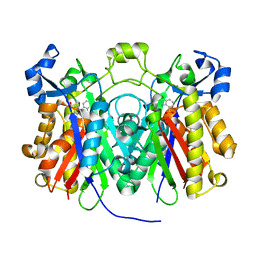 | | Crystal structure of Pseudomonas aeruginosa PqsD C112A mutant in complex with anthranilic acid | | Descriptor: | 2-AMINOBENZOIC ACID, PQS biosynthetic enzyme | | Authors: | Bera, A.K, Atanasova, V, Parsons, J.F. | | Deposit date: | 2009-04-24 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of PqsD, a Pseudomonas quinolone signal biosynthetic enzyme, in complex with anthranilate.
Biochemistry, 48, 2009
|
|
3H76
 
 | | Crystal structure of PqsD, a key enzyme in Pseudomonas aeruginosa quinolone signal biosynthesis pathway | | Descriptor: | PQS biosynthetic enzyme | | Authors: | Bera, A.K, Atanasova, V, Parsons, J.F. | | Deposit date: | 2009-04-24 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of PqsD, a Pseudomonas quinolone signal biosynthetic enzyme, in complex with anthranilate.
Biochemistry, 48, 2009
|
|
