1DNP
 
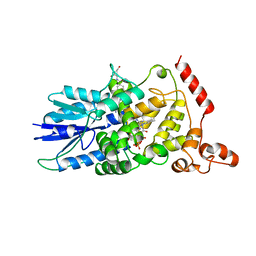 | | STRUCTURE OF DEOXYRIBODIPYRIMIDINE PHOTOLYASE | | Descriptor: | 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, DNA PHOTOLYASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Park, H.-W, Sancar, A, Deisenhofer, J. | | Deposit date: | 1995-07-03 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of DNA photolyase from Escherichia coli.
Science, 268, 1995
|
|
1QHT
 
 | |
4QX1
 
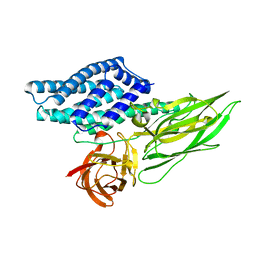 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the CrystFEL software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1OQN
 
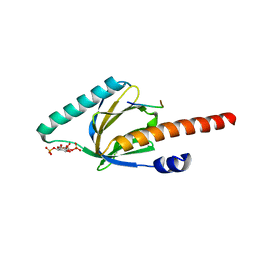 | | Crystal structure of the phosphotyrosine binding domain (PTB) of mouse Disabled 1 (Dab1) | | Descriptor: | Alzheimer's disease amyloid A4 protein homolog, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Disabled homolog 1 | | Authors: | Yun, M, Keshvara, L, Park, C.-G, Zhang, Y.-M, Dickerson, J.B, Zheng, J, Rock, C.O, Curran, T, Park, H.-W. | | Deposit date: | 2003-03-10 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the Dab homology domains of mouse disabled 1 and 2
J.Biol.Chem., 278, 2003
|
|
1M7E
 
 | | Crystal structure of the phosphotyrosine binding domain(PTB) of mouse Disabled 2(Dab2):implications for Reeling signaling | | Descriptor: | Disabled homolog 2, NGYENPTYK peptide | | Authors: | Yun, M, Keshvara, L, Park, C.-G, Zhang, Y.-M, Dickerson, J.B, Zheng, J, Rock, C.O, Curran, T, Park, H.-W. | | Deposit date: | 2002-07-19 | | Release date: | 2003-08-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of the Dab homology domains of mouse disabled 1 and 2
J.Biol.Chem., 278, 2003
|
|
4DVQ
 
 | | Structure of human aldosterone synthase, CYP11B2, in complex with deoxycorticosterone | | Descriptor: | Cytochrome P450 11B2, mitochondrial, DESOXYCORTICOSTERONE, ... | | Authors: | Strushkevich, N, Shen, L, Tempel, W, Arrowsmith, C, Edwards, A, Usanov, S.A, Park, H.-W. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insights into aldosterone synthase substrate specificity and targeted inhibition.
Mol.Endocrinol., 27, 2013
|
|
1F9T
 
 | | CRYSTAL STRUCTURES OF KINESIN MUTANTS REVEAL A SIGNALLING PATHWAY FOR ACTIVATION OF THE MOTOR ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KAR3, MAGNESIUM ION | | Authors: | Yun, M, Zhang, X, Park, C.-G, Park, H.-W, Endow, S.A. | | Deposit date: | 2000-07-11 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A structural pathway for activation of the kinesin motor ATPase.
EMBO J., 20, 2001
|
|
1N6M
 
 | | Rotation of the stalk/neck and one head in a new crystal structure of the kinesin motor protein, Ncd | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Claret segregational protein, MAGNESIUM ION | | Authors: | Yun, M, Bronner, C.E, Park, C.-G, Cha, S.-S, Park, H.-W, Endow, S.A. | | Deposit date: | 2002-11-11 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rotation of the stalk/neck and one head in a new crystal structure of the kinesin motor protein, Ncd
EMBO J., 22, 2003
|
|
4FDH
 
 | | Structure of human aldosterone synthase, CYP11B2, in complex with fadrozole | | Descriptor: | 4-[(5R)-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-5-yl]benzonitrile, Cytochrome P450 11B2, mitochondrial, ... | | Authors: | Strushkevich, N, Shen, L, Tempel, W, Arrowsmith, C, Edwards, A, Usanov, S.A, Park, H.-W. | | Deposit date: | 2012-05-28 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural insights into aldosterone synthase substrate specificity and targeted inhibition.
Mol.Endocrinol., 27, 2013
|
|
1DC5
 
 | |
1DC4
 
 | | STRUCTURAL ANALYSIS OF GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE FROM ESCHERICHIA COLI: DIRECT EVIDENCE FOR SUBSTRATE BINDING AND COFACTOR-INDUCED CONFORMATIONAL CHANGES | | Descriptor: | GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Yun, M, Park, C.-G, Kim, J.-Y, Park, H.-W. | | Deposit date: | 1999-11-04 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of glyceraldehyde 3-phosphate dehydrogenase from Escherichia coli: direct evidence of substrate binding and cofactor-induced conformational changes.
Biochemistry, 39, 2000
|
|
1DC6
 
 | | STRUCTURAL ANALYSIS OF GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE FROM ESCHERICHIA COLI: DIRECT EVIDENCE FOR SUBSTRATE BINDING AND COFACTOR-INDUCED CONFORMATIONAL CHANGES. | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yun, M, Park, C.-G, Kim, J.-Y, Park, H.-W. | | Deposit date: | 1999-11-04 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of glyceraldehyde 3-phosphate dehydrogenase from Escherichia coli: direct evidence of substrate binding and cofactor-induced conformational changes.
Biochemistry, 39, 2000
|
|
1FT1
 
 | |
4QX0
 
 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QX3
 
 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the CrystFEL software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QX2
 
 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1JYK
 
 | |
1JYL
 
 | |
1SQ5
 
 | | Crystal Structure of E. coli Pantothenate kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PANTOTHENOIC ACID, Pantothenate kinase | | Authors: | Ivey, R.A, Zhang, Y.-M, Virga, K.G, Hevener, K, Lee, R.E, Rock, C.O, Jackowski, S, Park, H.-W. | | Deposit date: | 2004-03-17 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the pantothenate kinase.ADP.pantothenate ternary complex reveals the relationship between the binding sites for substrate, allosteric regulator, and antimetabolites.
J.Biol.Chem., 279, 2004
|
|
