1YZK
 
 | | GppNHp bound Rab11 GTPase | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-11A | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-02-28 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1YZU
 
 | | GppNHp-Bound Rab21 GTPase at 2.50 A Resolution | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-21 | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-02-28 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1YZN
 
 | | GppNHp-Bound Ypt1p GTPase | | Descriptor: | GTP-binding protein YPT1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-02-28 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1Z0J
 
 | | Structure of GTP-Bound Rab22Q64L GTPase in complex with the minimal Rab binding domain of Rabenosyn-5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FYVE-finger-containing Rab5 effector protein rabenosyn-5, GLYCEROL, ... | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-03-01 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1Z07
 
 | | GppNHp-Bound Rab5c G55Q mutant GTPase | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-5C | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-03-01 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1Z2A
 
 | | GDP-Bound Rab23 GTPase crystallized in P2(1)2(1)2(1) space group | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-23 | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-03-07 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1Z0F
 
 | | GDP-Bound Rab14 GTPase | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAB14, ... | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-03-01 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1YZQ
 
 | | GppNHp-Bound Rab6 GTPase | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, small GTP binding protein RAB6 isoform | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-02-28 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1YZM
 
 | | Structure of Rabenosyn (458-503), Rab4 binding domain | | Descriptor: | FYVE-finger-containing Rab5 effector protein rabenosyn-5 | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-02-28 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1Z06
 
 | | GppNHp-Bound Rab33 GTPase | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-33B | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-03-01 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1Z0D
 
 | | GDP-Bound Rab5c GTPase | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-03-01 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1YZL
 
 | | GppNHp-Bound Rab9 GTPase | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-9A | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-02-28 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1Z0I
 
 | | GDP-Bound Rab21 GTPase | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-21 | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-03-01 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
4J1L
 
 | | Mutant Endotoxin TeNT | | Descriptor: | Tetanus toxin, ZINC ION | | Authors: | Guo, J, Pan, X, Zhao, Y, Chen, S. | | Deposit date: | 2013-02-01 | | Release date: | 2014-02-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering Clostridia Neurotoxins with elevated catalytic activity
Toxicon, 74, 2013
|
|
5VY1
 
 | | Crystal structure of the extended Tudor domain from BmPAPI | | Descriptor: | Tudor and KH domain-containing protein homolog | | Authors: | Hubbard, P.A, Pan, X, McNally, R, Ohtaki, A, Honda, S, Kirino, Y, Murali, R. | | Deposit date: | 2017-05-24 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural studies of the Tudor domain from the Bombyx homolog of Drosophila PAPI: Implication to piRNA biogenesis
To Be Published
|
|
5X0M
 
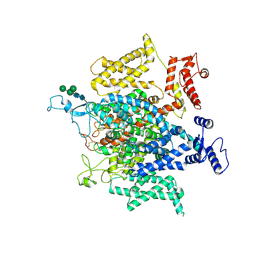 | | Structure of a eukaryotic voltage-gated sodium channel at near atomic resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein, ... | | Authors: | Shen, H, Zhou, Q, Pan, X, Li, Z, Wu, J, Yan, N. | | Deposit date: | 2017-01-21 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution.
Science, 355, 2017
|
|
5VQH
 
 | | Crystal structure of the extended Tudor domain from BmPAPI in complex with sDMA | | Descriptor: | N3, N4-DIMETHYLARGININE, Tudor and KH domain-containing protein homolog | | Authors: | Hubbard, P.A, Pan, X, Ohtaki, A, McNally, R, Honda, S, Kirino, Y, Murali, R. | | Deposit date: | 2017-05-08 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of the Tudor domain from the Bombyx homolog of Drosophila PAPI: Implication to piRNA biogenesis
To Be Published
|
|
5VQG
 
 | | Crystal structure of the extended Tudor domain from BmPAPI | | Descriptor: | Tudor and KH domain-containing protein homolog | | Authors: | Hubbard, P.A, Pan, X, Ohtaki, A, McNally, R, Honda, S, Kirino, Y, Murali, R. | | Deposit date: | 2017-05-08 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural studies of the Tudor domain from the Bombyx homolog of Drosophila PAPI: Implication to piRNA biogenesis
To Be Published
|
|
7DTC
 
 | | voltage-gated sodium channel Nav1.5-E1784K | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 5 subunit alpha | | Authors: | Yan, N, Pan, X, Li, Z. | | Deposit date: | 2021-01-04 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human Na v 1.5 reveals the fast inactivation-related segments as a mutational hotspot for the long QT syndrome.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DTD
 
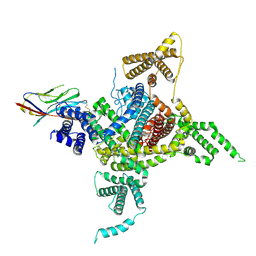 | | Voltage-gated sodium channel Nav1.1 and beta4 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 1 subunit alpha, ... | | Authors: | Yan, N, Pan, X, Li, Z, Huang, G. | | Deposit date: | 2021-01-04 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Comparative structural analysis of human Na v 1.1 and Na v 1.5 reveals mutational hotspots for sodium channelopathies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7XVE
 
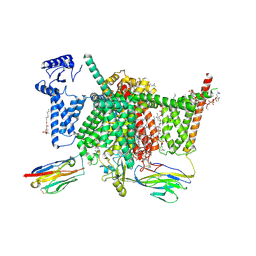 | | Human Nav1.7 mutant class-I | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Huang, G, Wu, Q, Li, Z, Pan, X, Yan, N. | | Deposit date: | 2022-05-21 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Unwinding and spiral sliding of S4 and domain rotation of VSD during the electromechanical coupling in Na v 1.7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XVF
 
 | | Nav1.7 mutant class2 | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Huang, G, Wu, Q, Li, Z, Pan, X, Yan, N. | | Deposit date: | 2022-05-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Unwinding and spiral sliding of S4 and domain rotation of VSD during the electromechanical coupling in Na v 1.7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XN6
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with ADPR-deacylization | | Descriptor: | Arginine ADP-riboxanase CopC, Calmodulin-1, Caspase-3, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
7XN4
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with NAD+ | | Descriptor: | Arginine ADP-riboxanase CopC, Calmodulin-1, Caspase-3, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
7XN5
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with ADPR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Arginine ADP-riboxanase CopC, Calmodulin-1, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
