2V68
 
 | | Crystal structure of Chlamydomonas reinhardtii Rubisco with large- subunit mutations V331A, T342I | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Satagopan, S, Taylor, T.C, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2007-07-13 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of Altered Large-Subunit Loop-6-Carboxy-Terminus Interactions that Influence Catalytic Efficiency and Co2 O2 Specificity of Ribulose-1,5-Bisphosphate Carboxylase Oxygenase
Biochemistry, 46, 2007
|
|
2V67
 
 | | Crystal structure of Chlamydomonas reinhardtii Rubisco with a large- subunit supressor mutation T342I | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Satagopan, S, Taylor, T.C, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2007-07-13 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Altered Large-Subunit Loop-6-Carboxy-Terminus Interactions that Influence Catalytic Efficiency and Co2 O2 Specificity of Ribulose-1,5-Bisphosphate Carboxylase Oxygenase
Biochemistry, 46, 2007
|
|
6PCA
 
 | | Crystal structure of beta-ketoadipyl-CoA thiolase | | Descriptor: | ACETATE ION, Beta-ketoadipyl-CoA thiolase, CHLORIDE ION, ... | | Authors: | Sukritee, B, Panjikar, S. | | Deposit date: | 2019-06-17 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for differentiation between two classes of thiolase: Degradative vs biosynthetic thiolase.
J Struct Biol X, 4, 2020
|
|
6W1J
 
 | | Cryo-EM structure of 5HT3A receptor in presence of Alosetron | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(4-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | Authors: | Basak, S, Chakrapani, S. | | Deposit date: | 2020-03-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | High-resolution structures of multiple 5-HT 3A R-setron complexes reveal a novel mechanism of competitive inhibition.
Elife, 9, 2020
|
|
2A7A
 
 | | On the Routine Use of Soft X-Rays in Macromolecular Crystallography, Part III- The Optimal Data Collection Wavelength | | Descriptor: | CALCIUM ION, Concanavalin A, MANGANESE (II) ION, ... | | Authors: | Mueller-Dieckmann, C, Panjikar, S, Tucker, P.A, Weiss, M.S. | | Deposit date: | 2005-07-05 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part III. The optimal data-collection wavelength.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2A7J
 
 | | On the Routine Use of Soft X-Rays in Macromolecular Crystallography, Part III- The Optimal Data Collection Wavelength | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Mueller-Dieckmann, C, Panjikar, S, Tucker, P.A, Weiss, M.S. | | Deposit date: | 2005-07-05 | | Release date: | 2005-07-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part III. The optimal data-collection wavelength.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2VDH
 
 | | Crystal structure of Chlamydomonas reinhardtii Rubisco with a large- subunit C172S mutation | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Garcia-Murria, M.-J, Karkehabadi, S, Marin-Navarro, J, Satagopan, S, Andersson, I, Spreitzer, R.J, Moreno, J. | | Deposit date: | 2007-10-09 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Consequences of the Replacement of Proximal Residues Cys-172 and Cys-192 in the Large Subunit of Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase from Chlamydomonas Reinhardtii
Biochem.J., 411, 2008
|
|
5IRA
 
 | | Expanding Nature's Catalytic Repertoire -Directed Evolution of an Artificial Metalloenzyme for In Vivo Metathesis | | Descriptor: | Artificial Metathesase, [1-[4-[[5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]pentanoylamino]methyl]-2,6-dimethyl-phenyl]-3-(2,4,6-trimethylphenyl)-4,5-dihydroimidazol-1-ium-2-yl]-bis(chloranyl)ruthenium | | Authors: | Heinisch, T, Jeschek, M, Reuter, R, Trindler, C, Panke, S, Ward, T.R. | | Deposit date: | 2016-03-12 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Directed evolution of artificial metalloenzymes for in vivo metathesis.
Nature, 537, 2016
|
|
6VM3
 
 | | Full length Glycine receptor reconstituted in lipid nanodisc in Gly/IVM-conformation (State-3) | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kumar, A, Basak, S, Chakrapani, S. | | Deposit date: | 2020-01-27 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Mechanisms of activation and desensitization of full-length glycine receptor in lipid nanodiscs.
Nat Commun, 11, 2020
|
|
1XWR
 
 | | Crystal structure of the coliphage lambda transcription activator protein CII | | Descriptor: | ISOPROPYL ALCOHOL, Regulatory protein CII | | Authors: | Datta, A.B, Panjikar, S, Weiss, M.S, Chakrabarti, P, Parrack, P. | | Deposit date: | 2004-11-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure of {lambda} CII: Implications for recognition of direct-repeat DNA by an unusual tetrameric organization
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6DG7
 
 | | Full-length 5-HT3A receptor in a serotonin-bound conformation- State 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, SEROTONIN, ... | | Authors: | Basak, S, Chakrapani, S. | | Deposit date: | 2018-05-17 | | Release date: | 2018-11-07 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM reveals two distinct serotonin-bound conformations of full-length 5-HT3Areceptor.
Nature, 563, 2018
|
|
2A7D
 
 | | On the Routine Use of Soft X-Rays in Macromolecular Crystallography, Part III- The Optimal Data Collection Wavelength | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Mueller-Dieckmann, C, Panjikar, S, Tucker, P.A, Weiss, M.S. | | Deposit date: | 2005-07-05 | | Release date: | 2005-07-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part III. The optimal data-collection wavelength.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2A7G
 
 | | On the Routine Use of Soft X-Rays in Macromolecular Crystallography, Part III- The Optimal Data Collection Wavelength | | Descriptor: | ACETIC ACID, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Mueller-Dieckmann, C, Panjikar, S, Tucker, P.A, Weiss, M.S. | | Deposit date: | 2005-07-05 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part III. The optimal data-collection wavelength.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2V69
 
 | | Crystal structure of Chlamydomonas reinhardtii Rubisco with a large- subunit mutation D473E | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Satagopan, S, Taylor, T.C, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2007-07-14 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis of Altered Large-Subunit Loop-6-Carboxy-Terminus Interactions that Influence Catalytic Efficiency and Co2-O2 Specificity of Ribulose-1,5-Bisphosphate Carboxylase Oxygenase
Biochemistry, 46, 2007
|
|
2A7B
 
 | | On the Routine Use of Soft X-Rays in Macromolecular Crystallography, Part III- The Optimal Data Collection Wavelength | | Descriptor: | XENON, gamma-adaptin appendage domain | | Authors: | Mueller-Dieckmann, C, Panjikar, S, Tucker, P.A, Weiss, M.S. | | Deposit date: | 2005-07-05 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part III. The optimal data-collection wavelength.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2A7I
 
 | | On the Routine Use of Soft X-Rays in Macromolecular Crystallography, Part III- The Optimal Data Collection Wavelength | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Mueller-Dieckmann, C, Panjikar, S, Tucker, P.A, Weiss, M.S. | | Deposit date: | 2005-07-05 | | Release date: | 2005-07-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part III. The optimal data-collection wavelength.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2A7H
 
 | | On the Routine Use of Soft X-Rays in Macromolecular Crystallography, Part III- The Optimal Data Collection Wavelength | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cationic trypsin | | Authors: | Mueller-Dieckmann, C, Panjikar, S, Tucker, P.A, Weiss, M.S. | | Deposit date: | 2005-07-05 | | Release date: | 2005-07-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part III. The optimal data-collection wavelength.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2V6A
 
 | | Crystal structure of Chlamydomonas reinhardtii Rubisco with large- subunit mutations V331A, G344S | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Satagopan, S, Taylor, T.C, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2007-07-14 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of Altered Large-Subunit Loop-6-Carboxy-Terminus Interactions that Influence Catalytic Efficiency and Co2-O2 Specificity of Ribulose-1,5-Bisphosphate Carboxylase Oxygenase
Biochemistry, 46, 2007
|
|
2VDI
 
 | | Crystal structure of Chlamydomonas reinhardtii Rubisco with a large- subunit C192S mutation | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Garcia-Murria, M.-J, Karkehabadi, S, Marin-Navarro, J, Satagopan, S, Andersson, I, Spreitzer, R.J, Moreno, J. | | Deposit date: | 2007-10-09 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Functional Consequences of the Replacement of Proximal Residues Cys-172 and Cys-192 in the Large Subunit of Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase from Chlamydomonas Reinhardtii
Biochem.J., 411, 2008
|
|
2A7C
 
 | | On the Routine Use of Soft X-Rays in Macromolecular Crystallography, Part III- The Optimal Data Collection Wavelength | | Descriptor: | Elastase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Mueller-Dieckmann, C, Panjikar, S, Tucker, P.A, Weiss, M.S. | | Deposit date: | 2005-07-05 | | Release date: | 2005-07-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part III. The optimal data-collection wavelength.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2A7F
 
 | | On the Routine Use of Soft X-Rays in Macromolecular Crystallography, Part III- The Optimal Data Collection Wavelength | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Mueller-Dieckmann, C, Panjikar, S, Tucker, P.A, Weiss, M.S. | | Deposit date: | 2005-07-05 | | Release date: | 2005-07-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part III. The optimal data-collection wavelength.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5J0Z
 
 | | Crystal structure of GLIC in complex with DHA | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Basak, S, Schmandt, N, Chakrapani, S. | | Deposit date: | 2016-03-28 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure and dynamics of a lipid-induced potential desensitized-state of a pentameric ligand-gated channel.
Elife, 6, 2017
|
|
3OC5
 
 | |
3OC8
 
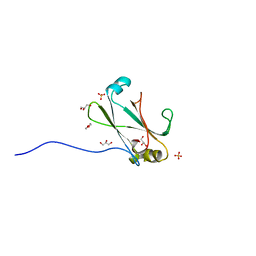 | | Crystal Structure of the C-terminal Domain of the Vibrio cholerae soluble colonization factor TcpF | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Craig, L, Kolappan, S, Yuen, A.S.W. | | Deposit date: | 2010-08-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Vibrio cholerae Colonization Factor TcpF and Identification of a Functional Immunogenic Site.
J.Mol.Biol., 409, 2011
|
|
2JKG
 
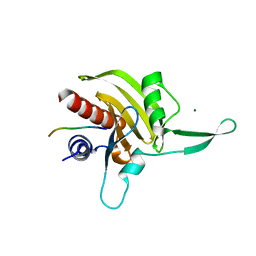 | | Plasmodium falciparum profilin | | Descriptor: | MAGNESIUM ION, OCTAPROLINE PEPTIDE, PROFILIN | | Authors: | Kursula, I, Kursula, P, Ganter, M, Panjikar, S, Matuschewski, K, Schueler, H. | | Deposit date: | 2008-08-28 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Basis for Parasite-Specific Functions of the Divergent Profilin of Plasmodium Falciparum.
Structure, 16, 2008
|
|
