5Y8X
 
 | | Crystal structure of Bacillus licheniformis Gamma glutamyl transpeptidase with Azaserine | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Kumari, S, Goel, M, Pal, R, Gupta, R. | | Deposit date: | 2017-08-21 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of Bacillus licheniformis Gamma glutamyl transpeptidase with Azaserine
To Be Published
|
|
7CLL
 
 | | Mycobacterium tubeculosis enolase in complex with 2-Phosphoglycerate | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, ACETATE ION, CHLORIDE ION, ... | | Authors: | Ahmad, M, Jha, B, Tiwari, S, Pal, R.K, Biswal, B.K. | | Deposit date: | 2020-07-21 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural snapshots of Mycobacterium tuberculosis enolase reveal dual mode of 2PG binding and its implication in enzyme catalysis.
Iucrj, 10, 2023
|
|
7Y52
 
 | | Crystal structure of peptidyl-tRNA hydrolase from Enterococcus faecium | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Peptidyl-tRNA hydrolase, ... | | Authors: | Pandey, R, Zohib, M, Mundra, S, Pal, R.K, Arora, A. | | Deposit date: | 2022-06-16 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from Enterococcus faecium
To Be Published
|
|
7CLK
 
 | | Mycobacterium tuberculosis enolase in complex with alternate 2-phosphoglycerate | | Descriptor: | 1,2-ETHANEDIOL, 2-PHOSPHOGLYCERIC ACID, ACETATE ION, ... | | Authors: | Ahmad, M, Jha, B, Tiwari, S, Pal, R.K, Biswal, B.K. | | Deposit date: | 2020-07-21 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural snapshots of Mycobacterium tuberculosis enolase reveal dual mode of 2PG binding and its implication in enzyme catalysis.
Iucrj, 10, 2023
|
|
7E4F
 
 | | Mycobacterium tuberculosis enolase mutant - E204A complex with phosphoenolpyruvate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ahmad, M, Pal, R.K, Biswal, B.K. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural snapshots of Mycobacterium tuberculosis enolase reveal dual mode of 2PG binding and its implication in enzyme catalysis.
Iucrj, 10, 2023
|
|
5ZQN
 
 | | Crystal structure of Mycobacterium tuberculosis HisB in complex with a ligand | | Descriptor: | (2R,3S)-2,3-dihydroxy-3-(1H-imidazol-5-yl)propyl dihydrogen phosphate, CHLORIDE ION, Imidazoleglycerol-phosphate dehydratase, ... | | Authors: | Kumar, D, Pal, R.K, Biswal, B.K. | | Deposit date: | 2018-04-19 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a triazole scaffold compound as an inhibitor of Mycobacterium tuberculosis imidazoleglycerol-phosphate dehydratase.
Proteins, 2021
|
|
7DDV
 
 | | Crystal structure of M.tuberculosis imidazole glycerol phosphate dehydratase in complex with an inhibitor | | Descriptor: | (1S)-1-(2-methyl-1,2,4-triazol-3-yl)ethanamine, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tiwari, S, Pal, R.K, Biswal, B.K. | | Deposit date: | 2020-10-29 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of M.tuberculosis imidazole glycerol phosphate dehydratase in complex with an inhibitor
To Be Published
|
|
7DNQ
 
 | | Crystal structure of M.tuberculosis imidazole glycerol phosphate dehydratase in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-(1H-1,2,4-triazol-5-ylsulfanyl)ethanamine, CHLORIDE ION, ... | | Authors: | Tiwari, S, Pal, R.K, Biswal, B.K. | | Deposit date: | 2020-12-10 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of M.tuberculosis imidazole glycerol phosphate dehydratase in complex with an inhibitor
To Be Published
|
|
7BRD
 
 | | Crystal structure of Peptidyl-tRNA hydrolase from Klebsiella pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Mundra, S, Zohib, M, Pal, R.K, Biswal, B.K, Arora, A. | | Deposit date: | 2020-03-27 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Structural and functional characterization of peptidyl-tRNA hydrolase from Klebsiella pneumoniae.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
6KHH
 
 | | Crystal Structure of HisB from Mycobacterium tuberculosis | | Descriptor: | ACETAMIDE, CHLORIDE ION, Imidazoleglycerol-phosphate dehydratase, ... | | Authors: | Kumar, D, Jha, B, Pal, R.K, Biswal, B.K. | | Deposit date: | 2019-07-15 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Characterization of a triazole scaffold compound as an inhibitor of Mycobacterium tuberculosis imidazoleglycerol-phosphate dehydratase.
Proteins, 2021
|
|
2I7A
 
 | | Domain IV of Human Calpain 13 | | Descriptor: | CALCIUM ION, Calpain 13, GLYCEROL, ... | | Authors: | Walker, J.R, Ng, K, Davis, T.L, Ravulapalli, R, Butler-cole, C, Finerty Jr, P.J, Newman, E.M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-30 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Human Calpain 13
To be Published
|
|
1JNV
 
 | | The Conformation of the Epsilon and Gamma Subunits within the E. coli F1 ATPase | | Descriptor: | ATP SYNTHASE ALPHA CHAIN, ATP SYNTHASE BETA CHAIN, ATP SYNTHASE EPSILON CHAIN, ... | | Authors: | Hausrath, A.C, Capaldi, R.A, Matthews, B.W. | | Deposit date: | 2001-07-25 | | Release date: | 2001-12-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | The conformation of the epsilon- and gamma-subunits within the Escherichia coli F(1) ATPase.
J.Biol.Chem., 276, 2001
|
|
7P4R
 
 | | Ultra High Resolution X-ray Structure of Orthorhombic Bovine Pancreatic Ribonuclease at 100K | | Descriptor: | ETHANOL, Ribonuclease pancreatic, SULFATE ION | | Authors: | Lisgarten, D.R, Palmer, R.A, Cooper, J.B, Naylor, C.E, Howlin, B.J, Lisgarten, J.N, Najmudin, S, Lobley, C.M.C. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Ultra-high resolution X-ray structure of orthorhombic bovine pancreatic Ribonuclease A at 100K.
BMC Chem, 17, 2023
|
|
6NB5
 
 | | Crystal structure of anti- MERS-CoV human neutralizing LCA60 antibody Fab fragment | | Descriptor: | LCA60 antigen-binding (Fab) fragment, heavy chain, light chain | | Authors: | Walls, A.J, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, J, Quispe, J, Cameroni, E, Gopal, R, Dai, M, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NB8
 
 | | Crystal structure of anti- SARS-CoV human neutralizing S230 antibody Fab fragment | | Descriptor: | S230 antigen-binding (Fab) fragment, heavy chain, light chain | | Authors: | Walls, A.J, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, J, Quispe, J, Cameroni, E, Gopal, R, Dai, M, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
4XZJ
 
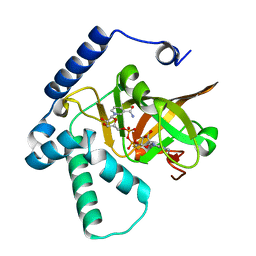 | | Crystal structure of ADP-ribosyltransferase Vis in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative NAD(+)--arginine ADP-ribosyltransferase Vis | | Authors: | Pfoh, R, Ravulapalli, R, Merrill, A.R, Pai, E.F. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of Vis Toxin, a Novel ADP-Ribosyltransferase from Vibrio splendidus.
Biochemistry, 54, 2015
|
|
4XZK
 
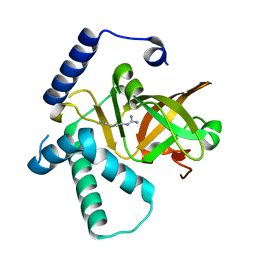 | |
4JFK
 
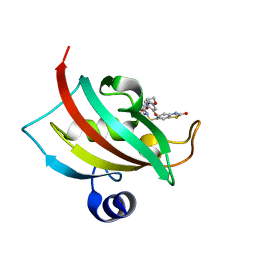 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with (1S,6R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-10-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,6R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-10-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4JFM
 
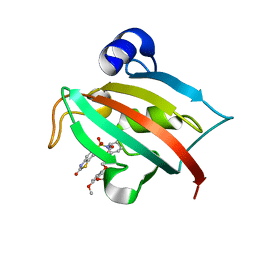 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with 2-(3,4-dimethoxyphenoxy)ethyl (2S)-1-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]piperidine-2-carboxylate | | Descriptor: | 2-(3,4-dimethoxyphenoxy)ethyl (2S)-1-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4JFL
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with 6-({(1S,5R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-2-oxo-3,9-diazabicyclo[3.3.1]non-9-yl}sulfonyl)-1,3-benzothiazol-2(3H)-one | | Descriptor: | 6-({(1S,5R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-2-oxo-3,9-diazabicyclo[3.3.1]non-9-yl}sulfonyl)-1,3-benzothiazol-2(3H)-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4JFI
 
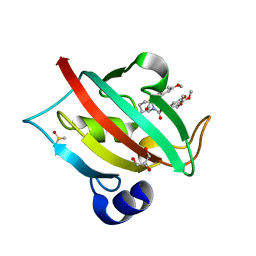 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with compound 1-[(9S,13R,13aR)-1,3-dimethoxy-8-oxo-5,8,9,10,11,12,13,13a-octahydro-6H-9,13-epiminoazocino[2,1-a]isoquinolin-14-yl]-2-(3,4,5-trimethoxyphenyl)ethane-1,2-dione | | Descriptor: | 1-[(9S,13R,13aR)-1,3-dimethoxy-8-oxo-5,8,9,10,11,12,13,13a-octahydro-6H-9,13-epiminoazocino[2,1-a]isoquinolin-14-yl]-2-(3,4,5-trimethoxyphenyl)ethane-1,2-dione, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4JFJ
 
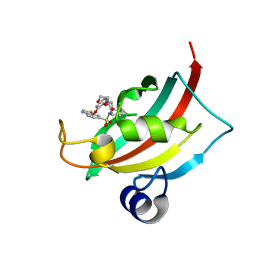 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with compound (1S,6R)-10-(1,3-benzothiazol-6-ylsulfonyl)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,6R)-10-(1,3-benzothiazol-6-ylsulfonyl)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
6NB4
 
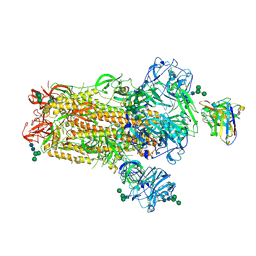 | | MERS-CoV S complex with human neutralizing LCA60 antibody Fab fragment (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LCA60 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NB7
 
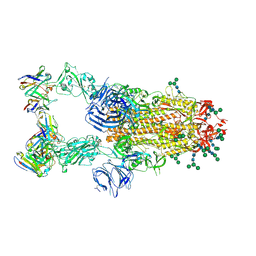 | | SARS-CoV complex with human neutralizing S230 antibody Fab fragment (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S230 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NB6
 
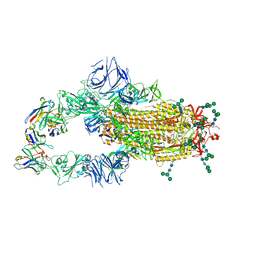 | | SARS-CoV complex with human neutralizing S230 antibody Fab fragment (state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S230 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
