1FMJ
 
 | | CRYSTAL STRUCTURE OF MERCURY DERIVATIVE OF RETINOL DEHYDRATASE IN A COMPLEX WITH RETINOL AND PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, CALCIUM ION, MERCURY (II) ION, ... | | Authors: | Pakhomova, S, Kobayashi, M, Buck, J, Newcomer, M.E. | | Deposit date: | 2000-08-17 | | Release date: | 2001-05-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A helical lid converts a sulfotransferase to a dehydratase.
Nat.Struct.Biol., 8, 2001
|
|
1FML
 
 | | CRYSTAL STRUCTURE OF RETINOL DEHYDRATASE IN A COMPLEX WITH RETINOL AND PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, CALCIUM ION, RETINOL, ... | | Authors: | Pakhomova, S, Kobayashi, M, Buck, J, Newcomer, M.E. | | Deposit date: | 2000-08-17 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A helical lid converts a sulfotransferase to a dehydratase.
Nat.Struct.Biol., 8, 2001
|
|
1NPB
 
 | | Crystal structure of the fosfomycin resistance protein from transposon Tn2921 | | Descriptor: | GLYCEROL, SULFATE ION, fosfomycin-resistance protein | | Authors: | Pakhomova, S, Rife, C.L, Armstrong, R.N, Newcomer, M.E. | | Deposit date: | 2003-01-17 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of fosfomycin resistance protein FosA from transposon Tn2921.
Protein Sci., 13, 2004
|
|
1X8L
 
 | | Crystal structure of retinol dehydratase in complex with all-trans-4-oxoretinol and inactive cofactor PAP | | Descriptor: | 4-OXORETINOL, ADENOSINE-3'-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Pakhomova, S, Buck, J, Newcomer, M.E. | | Deposit date: | 2004-08-18 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structures of the unique sulfotransferase retinol dehydratase with product and inhibitors provide insight into enzyme mechanism and inhibition.
Protein Sci., 14, 2005
|
|
1X8K
 
 | | Crystal structure of retinol dehydratase in complex with anhydroretinol and inactive cofactor PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, ANHYDRORETINOL, CALCIUM ION, ... | | Authors: | Pakhomova, S, Buck, J, Newcomer, M.E. | | Deposit date: | 2004-08-18 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structures of the unique sulfotransferase retinol dehydratase with product and inhibitors provide insight into enzyme mechanism and inhibition.
Protein Sci., 14, 2005
|
|
1X8J
 
 | | Crystal structure of retinol dehydratase in complex with androsterone and inactive cofactor PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Androsterone, CALCIUM ION, ... | | Authors: | Pakhomova, S, Buck, J, Newcomer, M.E. | | Deposit date: | 2004-08-18 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structures of the unique sulfotransferase retinol dehydratase with product and inhibitors provide insight into enzyme mechanism and inhibition.
Protein Sci., 14, 2005
|
|
3E4Y
 
 | |
3E4W
 
 | |
3D41
 
 | | Crystal structure of fosfomycin resistance kinase FomA from Streptomyces wedmorensis complexed with MgAMPPNP and fosfomycin | | Descriptor: | FOSFOMYCIN, FomA protein, MAGNESIUM ION, ... | | Authors: | Pakhomova, S, Bartlett, S.G, Augustus, A, Kuzuyama, T, Newcomer, M.E. | | Deposit date: | 2008-05-13 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Fosfomycin Resistance Kinase FomA from Streptomyces wedmorensis.
J.Biol.Chem., 283, 2008
|
|
3D40
 
 | | Crystal structure of fosfomycin resistance kinase FomA from Streptomyces wedmorensis complexed with diphosphate | | Descriptor: | DIPHOSPHATE, FomA protein | | Authors: | Pakhomova, S, Bartlett, S.G, Augustus, A, Kuzuyama, T, Newcomer, M.E. | | Deposit date: | 2008-05-13 | | Release date: | 2008-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal Structure of Fosfomycin Resistance Kinase FomA from Streptomyces wedmorensis.
J.Biol.Chem., 283, 2008
|
|
6NS6
 
 | | Crystal structure of fungal lipoxygenase from Fusarium graminearum. P21 crystal form. | | Descriptor: | FE (II) ION, lipoxygenase | | Authors: | Pakhomova, S, Boeglin, W.E, Neau, D.B, Bartlett, S.G, Brash, A.R, Newcomer, M.E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | An ensemble of lipoxygenase structures reveals novel conformations of the Fe coordination sphere.
Protein Sci., 28, 2019
|
|
6NS5
 
 | | Crystal structure of fungal lipoxygenase from Fusarium graminearum. Second C2 crystal form. | | Descriptor: | FE (II) ION, lipoxygenase | | Authors: | Pakhomova, S, Boeglin, W.E, Neau, D.B, Bartlett, S.G, Brash, A.R, Newcomer, M.E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | An ensemble of lipoxygenase structures reveals novel conformations of the Fe coordination sphere.
Protein Sci., 28, 2019
|
|
6NS2
 
 | | Crystal structure of fungal lipoxygenase from Fusarium graminearum. P212121 crystal form. | | Descriptor: | FE (II) ION, lipoxygenase | | Authors: | Pakhomova, S, Boeglin, W.E, Neau, D.B, Bartlett, S.G, Brash, A.R, Newcomer, M.E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | An ensemble of lipoxygenase structures reveals novel conformations of the Fe coordination sphere.
Protein Sci., 28, 2019
|
|
6NS4
 
 | | Crystal structure of fungal lipoxygenase from Fusarium graminearum. C2 crystal form. | | Descriptor: | ACETATE ION, FE (II) ION, GLYCEROL, ... | | Authors: | Pakhomova, S, Boeglin, W.E, Neau, D.B, Bartlett, S.G, Brash, A.R, Newcomer, M.E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An ensemble of lipoxygenase structures reveals novel conformations of the Fe coordination sphere.
Protein Sci., 28, 2019
|
|
6NS3
 
 | | Crystal structure of fungal lipoxygenase from Fusarium graminearum. I222 crystal form. | | Descriptor: | FE (II) ION, lipoxygenase | | Authors: | Pakhomova, S, Boeglin, W.E, Neau, D.B, Bartlett, S.G, Brash, A.R, Newcomer, M.E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | An ensemble of lipoxygenase structures reveals novel conformations of the Fe coordination sphere.
Protein Sci., 28, 2019
|
|
1R8X
 
 | | Crystal Structure of Mouse Glycine N-Methyltransferase (Tetragonal Form) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, glycine N-methyltransferase | | Authors: | Pakhomova, S, Luka, Z, Wagner, C, Newcomer, M.E. | | Deposit date: | 2003-10-28 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Glycine N-methyltransferases: a comparison of the crystal structures and kinetic properties of recombinant human, mouse and rat enzymes.
Proteins, 57, 2004
|
|
1R8Y
 
 | | Crystal Structure of Mouse Glycine N-Methyltransferase (Monoclinic Form) | | Descriptor: | BETA-MERCAPTOETHANOL, glycine N-methyltransferase | | Authors: | Pakhomova, S, Luka, Z, Wagner, C, Newcomer, M.E. | | Deposit date: | 2003-10-28 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Glycine N-methyltransferases: a comparison of the crystal structures and kinetic properties of recombinant human, mouse and rat enzymes.
Proteins, 57, 2004
|
|
1R74
 
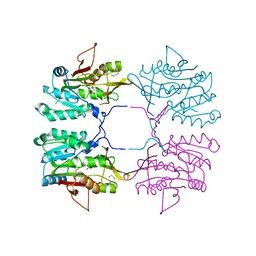 | | Crystal Structure of Human Glycine N-Methyltransferase | | Descriptor: | BETA-MERCAPTOETHANOL, CITRIC ACID, Glycine N-methyltransferase | | Authors: | Pakhomova, S, Luka, Z, Wagner, C, Newcomer, M.E. | | Deposit date: | 2003-10-17 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Glycine N-methyltransferases: a comparison of the crystal structures and kinetic properties of recombinant human, mouse and rat enzymes.
Proteins, 57, 2004
|
|
3QVF
 
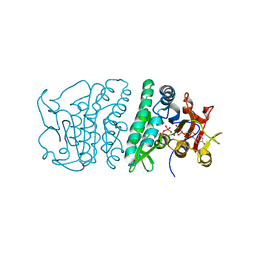 | | Crystal structure of fosfomycin resistance kinase FomA from Streptomyces wedmorensis complexed with MgADP and fosfomycin vanadate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FomA protein, MAGNESIUM ION, ... | | Authors: | Pakhomova, S, Bartlett, S.G, Doerner, P.A, Newcomer, M.E. | | Deposit date: | 2011-02-25 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biochemical insights into the mechanism of fosfomycin phosphorylation by fosfomycin resistance kinase FomA.
Biochemistry, 50, 2011
|
|
3QUN
 
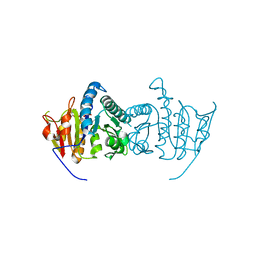 | | Crystal structure of fosfomycin resistance kinase FomA from Streptomyces wedmorensis complexed with MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FomA protein, GLYCEROL, ... | | Authors: | Pakhomova, S, Bartlett, S.G, Doerner, P.A, Newcomer, M.E. | | Deposit date: | 2011-02-24 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and biochemical insights into the mechanism of fosfomycin phosphorylation by fosfomycin resistance kinase FomA.
Biochemistry, 50, 2011
|
|
3QUO
 
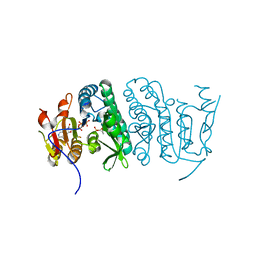 | | Crystal structure of fosfomycin resistance kinase FomA from Streptomyces wedmorensis complexed with ATP and fosfomycin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FOSFOMYCIN, FomA protein | | Authors: | Pakhomova, S, Bartlett, S.G, Doerner, P.A, Newcomer, M.E. | | Deposit date: | 2011-02-24 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural and biochemical insights into the mechanism of fosfomycin phosphorylation by fosfomycin resistance kinase FomA.
Biochemistry, 50, 2011
|
|
3QVH
 
 | | Crystal structure of fosfomycin resistance kinase FomA from Streptomyces wedmorensis complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FomA protein | | Authors: | Pakhomova, S, Bartlett, S.G, Doerner, P.A, Newcomer, M.E. | | Deposit date: | 2011-02-25 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biochemical insights into the mechanism of fosfomycin phosphorylation by fosfomycin resistance kinase FomA.
Biochemistry, 50, 2011
|
|
3QUR
 
 | | Crystal structure of fosfomycin resistance kinase FomA from Streptomyces wedmorensis complexed with MgADP and fosfomycin monophosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FomA protein, MAGNESIUM ION, ... | | Authors: | Pakhomova, S, Bartlett, S.G, Doerner, P.A, Newcomer, M.E. | | Deposit date: | 2011-02-24 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural and biochemical insights into the mechanism of fosfomycin phosphorylation by fosfomycin resistance kinase FomA.
Biochemistry, 50, 2011
|
|
4MV7
 
 | | Crystal Structure of Biotin Carboxylase form Haemophilus influenzae in Complex with Phosphonoformate | | Descriptor: | 1,2-ETHANEDIOL, Biotin carboxylase, PHOSPHONOFORMIC ACID | | Authors: | Broussard, T.C, Pakhomova, S, Neau, D.B, Champion, T.S, Bonnot, R.J, Waldrop, G.L. | | Deposit date: | 2013-09-23 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
Biochemistry, 54, 2015
|
|
4MV6
 
 | | Crystal Structure of Biotin Carboxylase from Haemophilus influenzae in Complex with Phosphonoacetamide | | Descriptor: | 1,2-ETHANEDIOL, Biotin carboxylase, PHOSPHONOACETAMIDE | | Authors: | Broussard, T.C, Pakhomova, S, Neau, D.B, Champion, T.S, Bonnot, R.J, Waldrop, G.L. | | Deposit date: | 2013-09-23 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
Biochemistry, 54, 2015
|
|
