2JOB
 
 | | Solution structure of an antilipopolysaccharide factor from shrimp and its possible Lipid A binding site | | Descriptor: | antilipopolysaccharide factor | | Authors: | Yang, Y, Boze, H, Chemardin, P, Padilla, A, Moulin, G, Tassanakajon, A, Pugniere, M, Roquet, F, Gueguen, Y, Bachere, E, Aumelas, A. | | Deposit date: | 2007-03-02 | | Release date: | 2008-03-11 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR structure of rALF-Pm3, an anti-lipopolysaccharide factor from shrimp: Model of the possible lipid A-binding site
Biopolymers, 91, 2009
|
|
6R5J
 
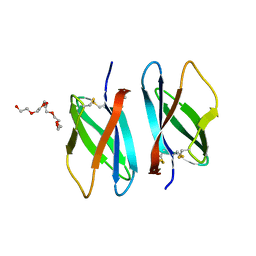 | |
1R3B
 
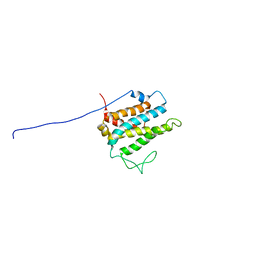 | | Solution structure of xenopus laevis Mob1 | | Descriptor: | MOB1 | | Authors: | Ponchon, L, Dumas, C, Kajava, A.V, Fesquet, D, Padilla, A. | | Deposit date: | 2003-10-01 | | Release date: | 2004-09-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of Mob1, a mitotic exit network protein and its interaction with an NDR kinase peptide
J.Mol.Biol., 337, 2004
|
|
4FYI
 
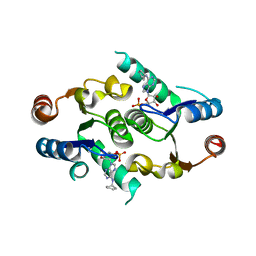 | | Crystal structure of rcl with 6-cyclopentyl-AMP | | Descriptor: | Deoxyribonucleoside 5'-monophosphate N-glycosidase, N-cyclopentyladenosine 5'-(dihydrogen phosphate) | | Authors: | Labesse, G, Padilla, A, Kaminski, P.A. | | Deposit date: | 2012-07-04 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of the oncoprotein Rcl bound to three nucleotide analogues.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
