4OD1
 
 | | Crystal structure of human Fab CAP256-VRC26.03, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.03 heavy chain, CAP256-VRC26.03 light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4OGV
 
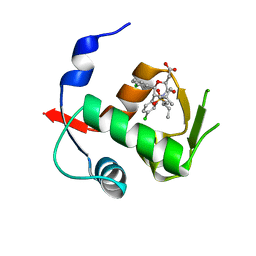 | | Co-Crystal Structure of MDM2 with Inhibitor Compound 49 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, [(2S,5R,6R)-4-[(2S)-1-(tert-butylsulfonyl)butan-2-yl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-3-oxomorpholin-2-yl]acetic acid | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
4OGN
 
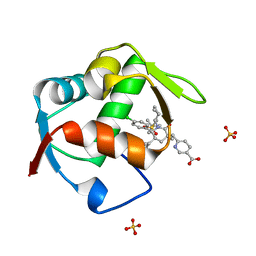 | | Co-Crystal Structure of MDM2 with Inhbitor Compound 3 | | Descriptor: | 6-{[(3R,5R,6S)-1-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-5-(3-chlorophenyl)-6-(4-chlorophenyl)-3-methyl-2-oxopiperidin-3-yl]methyl}pyridine-3-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.377 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
4OGT
 
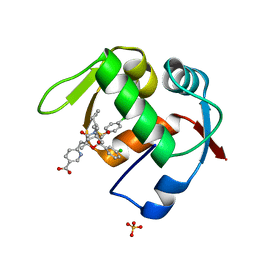 | | Co-Crystal Structure of MDM2 with Inhbitor Compound 46 | | Descriptor: | 6-{[(2R,5R,6R)-4-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-2-methyl-3-oxomorpholin-2-yl]methyl}pyridine-3-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5361 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
4O8V
 
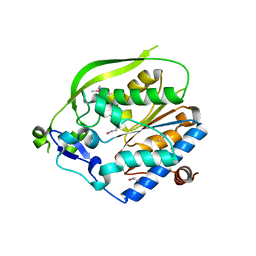 | | O-Acetyltransferase Domain of Pseudomonas putida AlgJ | | Descriptor: | Alginate biosynthesis protein AlgJ | | Authors: | Ricer, T, Little, D.J, Whitney, J.C, Robinson, H, Howell, P.L. | | Deposit date: | 2013-12-30 | | Release date: | 2014-10-01 | | Method: | X-RAY DIFFRACTION (1.815 Å) | | Cite: | P. aeruginosa SGNH Hydrolase-Like Proteins AlgJ and AlgX Have Similar Topology but Separate and Distinct Roles in Alginate Acetylation.
Plos Pathog., 10, 2014
|
|
4OBA
 
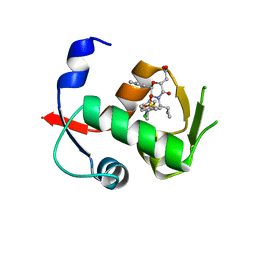 | | Co-crystal structure of MDM2 with Inhibitor Compound 4 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, [(2R,5R,6R)-4-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-3-oxomorpholin-2-yl]acetic acid | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-07 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Selective and Potent Morpholinone Inhibitors of the MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 57, 2014
|
|
4OCR
 
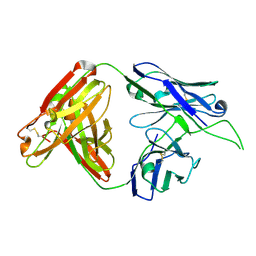 | | Crystal structure of human Fab CAP256-VRC26.01, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.01 heavy chain, CAP256-VRC26.01 light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4ODF
 
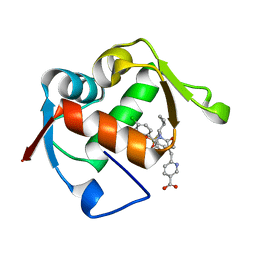 | | Co-Crystal Structure of MDM2 with Inhibitor Compound 47 | | Descriptor: | 6-{[(2S,5R,6R)-4-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-2-methyl-3-oxomorpholin-2-yl]methyl}pyridine-3-carboxylic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2006 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
4NK8
 
 | |
4P7O
 
 | | Structure of Escherichia coli PgaB C-terminal domain, P1 crystal form | | Descriptor: | Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P99
 
 | | Ca2+-stabilized adhesin helps an Antarctic bacterium reach out and bind ice | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Guo, S, Vance, D.R.T, Campbell, R.L, Davies, P.L. | | Deposit date: | 2014-04-02 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ca2+-stabilized adhesin helps an Antarctic bacterium reach out and bind ice.
Biosci.Rep., 34, 2014
|
|
4P7Q
 
 | | Structure of Escherichia coli PgaB C-terminal domain in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P7R
 
 | | Structure of Escherichia coli PgaB C-terminal domain in complex with a poly-beta-1,6-N-acetyl-D-glucosamine (PNAG) hexamer | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P7N
 
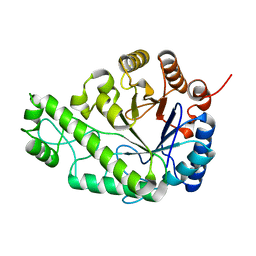 | | Structure of Escherichia coli PgaB C-terminal domain in complex with glucosamine | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1HY1
 
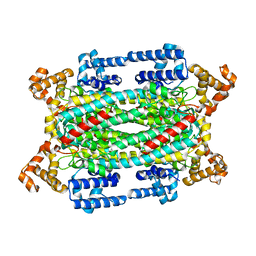 | |
1I0A
 
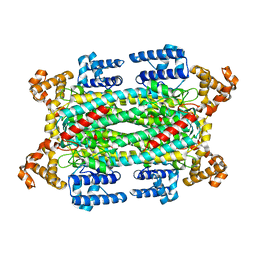 | |
1JAB
 
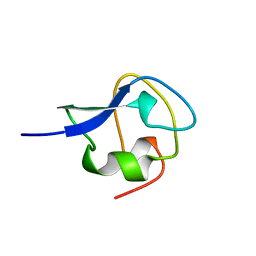 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T18S | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1K62
 
 | |
1KB4
 
 | |
1KB6
 
 | |
1KB2
 
 | |
1K97
 
 | |
1KKT
 
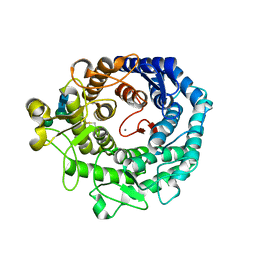 | | Structure of P. citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the ER and Golgi Class I enzymes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Mannosyl-oligosaccharide alpha-1,2-mannosidase, ... | | Authors: | Lobsanov, Y.D, Vallee, F, Imberty, A, Yoshida, T, Yip, P, Herscovics, A, Howell, P.L. | | Deposit date: | 2001-12-10 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Penicillium citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the endoplasmic reticulum and Golgi class I enzymes.
J.Biol.Chem., 277, 2002
|
|
1I6G
 
 | |
1KP2
 
 | |
