1GW6
 
 | | STRUCTURE OF LEUKOTRIENE A4 HYDROLASE D375N MUTANT | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, ACETATE ION, IMIDAZOLE, ... | | Authors: | Rudberg, P.C, Tholander, F, Thunnissen, M.M.G.M, Samuelsson, B, Haeggstrom, J.Z. | | Deposit date: | 2002-03-07 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Leukotriene A4 Hydrolase: Selective Abrogation of Leukotriene B4 Formation by Mutation of Aspartic Acid 375
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GVS
 
 | |
1H51
 
 | | Oxidised Pentaerythritol Tetranitrate Reductase (SCN complex) | | Descriptor: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE, THIOCYANATE ION | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2001-05-17 | | Release date: | 2003-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
1PNJ
 
 | | SOLUTION STRUCTURE AND LIGAND-BINDING SITE OF THE SH3 DOMAIN OF THE P85ALPHA SUBUNIT OF PHOSPHATIDYLINOSITOL 3-KINASE | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN | | Authors: | Booker, G.W, Gout, I, Downing, A.K, Driscoll, P.C, Boyd, J, Waterfield, M.D, Campbell, I.D. | | Deposit date: | 1993-07-19 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and ligand-binding site of the SH3 domain of the p85 alpha subunit of phosphatidylinositol 3-kinase.
Cell(Cambridge,Mass.), 73, 1993
|
|
1PNM
 
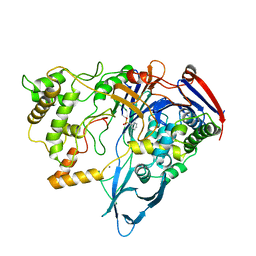 | |
1G2O
 
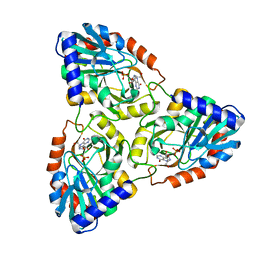 | | CRYSTAL STRUCTURE OF PURINE NUCLEOSIDE PHOSPHORYLASE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH A TRANSITION-STATE INHIBITOR | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PHOSPHATE ION, PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Shi, W, Basso, L.A, Tyler, P.C, Furneaux, R.H, Blanchard, J.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2000-10-20 | | Release date: | 2001-08-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of purine nucleoside phosphorylase from Mycobacterium tuberculosis in complexes with immucillin-H and its pieces.
Biochemistry, 40, 2001
|
|
1GAM
 
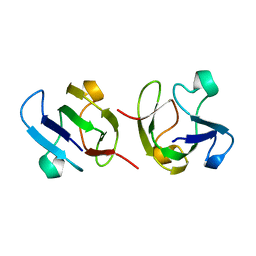 | | GAMMA B CRYSTALLIN TRUNCATED C-TERMINAL DOMAIN | | Descriptor: | GAMMA B CRYSTALLIN | | Authors: | Norledge, B.V, Mayr, E.-M, Glockshuber, R, Bateman, O.A, Slingsby, C, Jaenicke, R, Driessen, H.P.C. | | Deposit date: | 1996-02-02 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The X-ray structures of two mutant crystallin domains shed light on the evolution of multi-domain proteins.
Nat.Struct.Biol., 3, 1996
|
|
1GGE
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, NATIVE STRUCTURE AT 1.9 A RESOLUTION. | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, PROTEIN (CATALASE HPII) | | Authors: | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | Deposit date: | 2000-08-16 | | Release date: | 2000-08-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
1GGJ
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, ASN201ALA VARIANT. | | Descriptor: | CATALASE HPII, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE | | Authors: | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | Deposit date: | 2000-08-21 | | Release date: | 2000-08-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
1QLW
 
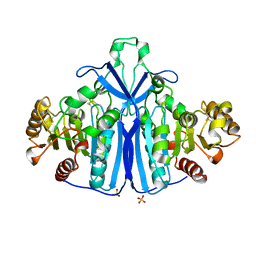 | |
1Q2Z
 
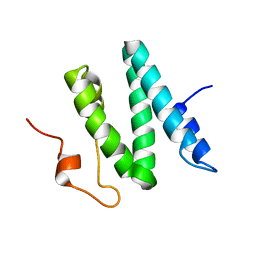 | | The 3D solution structure of the C-terminal region of Ku86 | | Descriptor: | ATP-dependent DNA helicase II, 80 kDa subunit | | Authors: | Harris, R, Esposito, D, Sankar, A, Maman, J.D, Hinks, J.A, Pearl, L.H, Driscoll, P.C. | | Deposit date: | 2003-07-28 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The 3D Solution Structure of the C-terminal Region of Ku86 (Ku86CTR)
J.Mol.Biol., 335, 2004
|
|
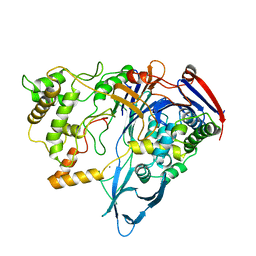
1PNK
 
 | |
1GQ0
 
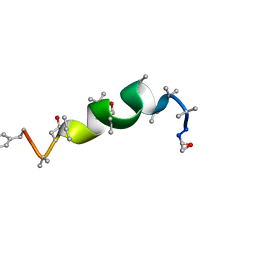 | | Solution structure of Antiamoebin I, a membrane channel-forming polypeptide; NMR, 20 structures | | Descriptor: | ANTIAMOEBIN I | | Authors: | Galbraith, T.P, Harris, R, Driscoll, P.C, Wallace, B.A. | | Deposit date: | 2001-11-16 | | Release date: | 2003-01-24 | | Last modified: | 2017-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution NMR studies of antiamoebin, a membrane channel-forming polypeptide.
Biophys. J., 84, 2003
|
|
1H61
 
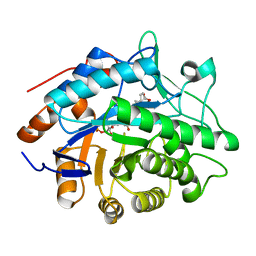 | | Structure of Pentaerythritol Tetranitrate Reductase in complex with prednisone | | Descriptor: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T.M, Moody, P.C.E. | | Deposit date: | 2001-06-04 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
1GVR
 
 | | STRUCTURE OF PENTAERYTHRITOL TETRANITRATE REDUCTASE AND COMPLEXED WITH 2,4,6 TRINITROTOLUENE | | Descriptor: | 2,4,6-TRINITROTOLUENE, FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2002-02-27 | | Release date: | 2003-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Kinetic and Structural Basis of Reactivity of Pentaerythritol Tetranitrate Reductase with Nadph,2-Cyclohexenone Nitroesters and Nitroaromatic Explosives
J.Biol.Chem., 277, 2002
|
|
1QWS
 
 | | Structure of the D181N variant of catalase HPII from E. coli | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chelikani, P, Carpena, X, Fita, I, Loewen, P.C. | | Deposit date: | 2003-09-03 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An electrical potential in the access channel of catalases enhances catalysis
J.Biol.Chem., 278, 2003
|
|
1H50
 
 | | Structure of Pentaerythritol Tetranitrate Reductase and complexes | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2001-05-17 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
1H1K
 
 | | THE BLUETONGUE VIRUS (BTV) CORE BINDS DSRNA | | Descriptor: | RNA | | Authors: | Diprose, J.M, Grimes, J.M, Sutton, G.C, Burroughs, J.N, Meyer, A, Maan, S, Mertens, P.P.C, Stuart, D.I. | | Deposit date: | 2002-07-17 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (10 Å) | | Cite: | The Core of Bluetongue Virus Binds Double-Stranded RNA
J.Virol., 76, 2002
|
|
1F2Z
 
 | |
1F35
 
 | |
1GVO
 
 | | STRUCTURE OF PENTAERYTHRITOL TETRANITRATE REDUCTASE AND COMPLEXED WITH 2,4 DINITROPHENOL | | Descriptor: | 2,4-DINITROPHENOL, FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2002-02-22 | | Release date: | 2003-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Kinetic and Structural Basis of Reactivity of Pentaerythritol Tetranitrate Reductase with Nadph,2-Cyclohexenone Nitroesters and Nitroaromatic Explosives
J.Biol.Chem., 277, 2002
|
|
1NHN
 
 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
NUCLEIC ACIDS MOL.BIOL., 9, 1995
|
|
1DQP
 
 | | CRYSTAL STRUCTURE OF GIARDIA GUANINE PHOSPHORIBOSYLTRANSFERASE COMPLEXED WITH IMMUCILLING | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-1-(S)-(9-DEAZAGUANIN-9-YL)-D-RIBITOL, GUANINE PHOSPHORIBOSYLTRANSFERASE, ISOPROPYL ALCOHOL | | Authors: | Shi, W, Munagala, N.R, Wang, C.C, Li, C.M, Tyler, P.C, Furneaux, R.H, Grubmeyer, C, Schramm, V.L, Almo, S.C. | | Deposit date: | 2000-01-04 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Giardia lamblia guanine phosphoribosyltransferase at 1.75 A(,).
Biochemistry, 39, 2000
|
|
1NHM
 
 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
NUCLEIC ACIDS MOL.BIOL., 9, 1995
|
|
1GOL
 
 | |
