4QON
 
 | | Structure of Bacillus pumilus catalase with catechol bound. | | Descriptor: | CATECHOL, CHLORIDE ION, Catalase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
5GI6
 
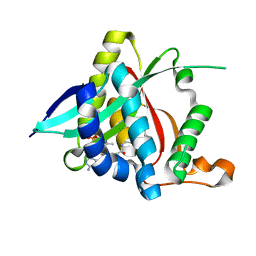 | |
4QOR
 
 | | Structure of Bacillus pumilus catalase with chlorophenol bound. | | Descriptor: | 2-CHLOROPHENOL, CHLORIDE ION, Catalase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
5GIG
 
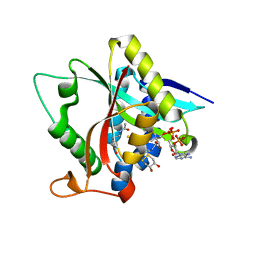 | | Crystal Structure of Drosophila melanogaster E47D Dopamine N-Acetyltransferase in Ternary Complex with CoA and Acetyl-dopamine | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, COENZYME A, Dopamine N-acetyltransferase, ... | | Authors: | Yang, Y.C, Wu, C.Y, Cheng, H.C, Lyu, P.C. | | Deposit date: | 2016-06-23 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Drosophila melanogaster E47D Dopamine N-Acetyltransferase in Ternary Complex with CoA and Acetyl-dopamine
To Be Published
|
|
5GM8
 
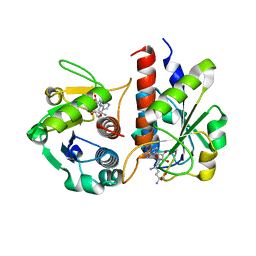 | | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruiginosa | | Descriptor: | SINEFUNGIN, tRNA (cytidine/uridine-2'-O-)-methyltransferase TrmJ | | Authors: | Jaroensuk, J, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Liew, C.W, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2016-07-13 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruginosa.
Nucleic Acids Res., 44, 2016
|
|
5GMC
 
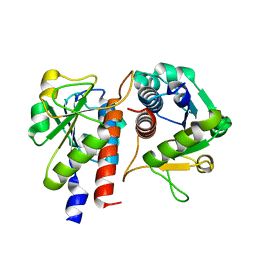 | | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruiginosa | | Descriptor: | tRNA (cytidine/uridine-2'-O-)-methyltransferase TrmJ | | Authors: | Jaroensuk, J, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Liew, C.W, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2016-07-13 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruginosa.
Nucleic Acids Res., 44, 2016
|
|
3H7W
 
 | | Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains with the artificial ligand THS017 | | Descriptor: | 2-nitro-N-(thiophen-3-ylmethyl)-4-(trifluoromethyl)aniline, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Key, J.M, Scheuermann, T.H, Anderson, P.C, Daggett, V, Gardner, K.H. | | Deposit date: | 2009-04-28 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Principles of ligand binding within a completely buried cavity in HIF2alpha PAS-B
J.Am.Chem.Soc., 131, 2009
|
|
3H82
 
 | | Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains with the artificial ligand THS020 | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, N-(furan-2-ylmethyl)-2-nitro-4-(trifluoromethyl)aniline | | Authors: | Key, J.M, Scheuermann, T.H, Anderson, P.C, Daggett, V, Gardner, K.H. | | Deposit date: | 2009-04-28 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Principles of ligand binding within a completely buried cavity in HIF2alpha PAS-B
J.Am.Chem.Soc., 131, 2009
|
|
4IJC
 
 | | Crystal structure of arabinose dehydrogenase Ara1 from Saccharomyces cerevisiae | | Descriptor: | D-arabinose dehydrogenase [NAD(P)+] heavy chain, GLYCEROL, SULFATE ION | | Authors: | Hu, X.Q, Guo, P.C, Li, W.F, Zhou, C.Z. | | Deposit date: | 2012-12-21 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Saccharomyces cerevisiaeD-arabinose dehydrogenase Ara1 and its complex with NADPH: implications for cofactor-assisted substrate recognition
Acta Crystallogr.,Sect.F, 69, 2013
|
|
5EKL
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS T62D/N100E at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Skerritt, L.A, Bell-Upp, P.C, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2015-11-03 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS T62D/N100E at cryogenic temperature
To be Published
|
|
3HO8
 
 | |
4Q3G
 
 | | Structure of the OsSERK2 leucine rich repeat extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, OsSERK2 | | Authors: | McAndrew, R.P, Pruitt, R.N, Kamita, S.G, Pereira, J.H, Majumder, D, Hammock, B.D, Adams, P.D, Ronald, P.C. | | Deposit date: | 2014-04-11 | | Release date: | 2014-11-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.787 Å) | | Cite: | Structure of the OsSERK2 leucine-rich repeat extracellular domain.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4IJR
 
 | | Crystal structure of Saccharomyces cerevisiae arabinose dehydrogenase Ara1 complexed with NADPH | | Descriptor: | D-arabinose dehydrogenase [NAD(P)+] heavy chain, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hu, X.Q, Guo, P.C, Li, W.F, Zhou, C.Z. | | Deposit date: | 2012-12-23 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Saccharomyces cerevisiaeD-arabinose dehydrogenase Ara1 and its complex with NADPH: implications for cofactor-assisted substrate recognition
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4G41
 
 | | Crystal structure of s-adenosylhomocysteine nucleosidase from streptococcus pyogenes in complex with 5-methylthiotubericidin | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MTA/SAH nucleosidase | | Authors: | Ponniah, K, Norris, G.E, Anderson, B.F, Brown, R.L, Tyler, P.C, Evans, G.B, Frohlich, R. | | Deposit date: | 2012-07-15 | | Release date: | 2012-09-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of s-adenosylhomocysteine nucleosidase from streptococcus pyogenes in complex with 5-methylthiotubericidin;
TO BE PUBLISHED
|
|
4QOS
 
 | | CRYSTAL STRUCTURE OF PSPF(1-265) E108Q MUTANT bound to ADP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | Deposit date: | 2014-06-20 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
4QNR
 
 | | CRYSTAL STRUCTURE OF PSPF(1-265) E108Q MUTANT bound to ATP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
4IZ8
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS H8E at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Bell-Upp, P.C, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-01-29 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS H8E at cryogenic temperature
To be Published
|
|
4J1M
 
 | |
4QNM
 
 | | CRYSTAL STRUCTURE of PSPF(1-265) E108Q MUTANT | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Psp operon transcriptional activator | | Authors: | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.628 Å) | | Cite: | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
2BYF
 
 | | NMR solution structure of phospholipase c epsilon RA 2 domain | | Descriptor: | PHOSPHOLIPASE C, EPSILON 1 | | Authors: | Bunney, T.D, Harris, R, Gandarillas, N.L, Josephs, M.B, Roe, S.M, Paterson, H.F, Rodrigues-Lima, F, Esposito, D, Gieschik, P, Pearl, L.H, Driscoll, P.C, Katan, M. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Mechanistic Insights Into Ras Association Domains of Phospholipase C Epsilon.
Mol.Cell, 21, 2006
|
|
2BYE
 
 | | NMR solution structure of phospholipase c epsilon RA 1 domain | | Descriptor: | PHOSPHOLIPASE C, EPSILON 1 | | Authors: | Bunney, T.D, Harris, R, Gandarillas, N.L, Josephs, M.B, Roe, S.M, Paterson, H.F, Rodrigues-Lima, F, Esposito, D, Gieschik, P, Pearl, L.H, Driscoll, P.C, Katan, M. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Mechanistic Insights Into Ras Association Domains of Phospholipase C Epsilon.
Mol.Cell, 21, 2006
|
|
2BGT
 
 | |
2BGU
 
 | | CRYSTAL STRUCTURE OF THE DNA MODIFYING ENZYME BETA-GLUCOSYLTRANSFERASE IN THE PRESENCE AND ABSENCE OF THE SUBSTRATE URIDINE DIPHOSPHOGLUCOSE | | Descriptor: | BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Vrielink, A, Rueger, W, Driessen, H.P.C, Freemont, P.S. | | Deposit date: | 1994-06-09 | | Release date: | 1995-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the DNA modifying enzyme beta-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose.
EMBO J., 13, 1994
|
|
2BTV
 
 | | ATOMIC MODEL FOR BLUETONGUE VIRUS (BTV) CORE | | Descriptor: | PROTEIN (VP3 CORE PROTEIN), PROTEIN (VP7 CORE PROTEIN) | | Authors: | Grimes, J.M, Burroughs, J.N, Gouet, P, Diprose, J.M, Malby, R, Zientras, S, Mertens, P.P.C, Stuart, D.I. | | Deposit date: | 1998-09-05 | | Release date: | 1998-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The atomic structure of the bluetongue virus core.
Nature, 395, 1998
|
|
2BLM
 
 | |
