1DQP
 
 | | CRYSTAL STRUCTURE OF GIARDIA GUANINE PHOSPHORIBOSYLTRANSFERASE COMPLEXED WITH IMMUCILLING | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-1-(S)-(9-DEAZAGUANIN-9-YL)-D-RIBITOL, GUANINE PHOSPHORIBOSYLTRANSFERASE, ISOPROPYL ALCOHOL | | Authors: | Shi, W, Munagala, N.R, Wang, C.C, Li, C.M, Tyler, P.C, Furneaux, R.H, Grubmeyer, C, Schramm, V.L, Almo, S.C. | | Deposit date: | 2000-01-04 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Giardia lamblia guanine phosphoribosyltransferase at 1.75 A(,).
Biochemistry, 39, 2000
|
|
1GQ0
 
 | | Solution structure of Antiamoebin I, a membrane channel-forming polypeptide; NMR, 20 structures | | Descriptor: | ANTIAMOEBIN I | | Authors: | Galbraith, T.P, Harris, R, Driscoll, P.C, Wallace, B.A. | | Deposit date: | 2001-11-16 | | Release date: | 2003-01-24 | | Last modified: | 2017-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution NMR studies of antiamoebin, a membrane channel-forming polypeptide.
Biophys. J., 84, 2003
|
|
1H61
 
 | | Structure of Pentaerythritol Tetranitrate Reductase in complex with prednisone | | Descriptor: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T.M, Moody, P.C.E. | | Deposit date: | 2001-06-04 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
1GVR
 
 | | STRUCTURE OF PENTAERYTHRITOL TETRANITRATE REDUCTASE AND COMPLEXED WITH 2,4,6 TRINITROTOLUENE | | Descriptor: | 2,4,6-TRINITROTOLUENE, FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2002-02-27 | | Release date: | 2003-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Kinetic and Structural Basis of Reactivity of Pentaerythritol Tetranitrate Reductase with Nadph,2-Cyclohexenone Nitroesters and Nitroaromatic Explosives
J.Biol.Chem., 277, 2002
|
|
1H50
 
 | | Structure of Pentaerythritol Tetranitrate Reductase and complexes | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2001-05-17 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
1H1K
 
 | | THE BLUETONGUE VIRUS (BTV) CORE BINDS DSRNA | | Descriptor: | RNA | | Authors: | Diprose, J.M, Grimes, J.M, Sutton, G.C, Burroughs, J.N, Meyer, A, Maan, S, Mertens, P.P.C, Stuart, D.I. | | Deposit date: | 2002-07-17 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (10 Å) | | Cite: | The Core of Bluetongue Virus Binds Double-Stranded RNA
J.Virol., 76, 2002
|
|
1F2E
 
 | | STRUCTURE OF SPHINGOMONAD, GLUTATHIONE S-TRANSFERASE COMPLEXED WITH GLUTATHIONE | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Nishio, T, Watanabe, T, Patel, A, Wang, Y, Lau, P.C.K, Grochulski, P, Li, Y, Cygler, M. | | Deposit date: | 2000-05-24 | | Release date: | 2000-06-21 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Properties of a Sphingomonad and Marine Bacterium Beta-Class Glutathione S-Transferases and Crystal Structure of the Former Complex with Glutathione
To be published
|
|
1F2M
 
 | |
1F2Y
 
 | |
1GVO
 
 | | STRUCTURE OF PENTAERYTHRITOL TETRANITRATE REDUCTASE AND COMPLEXED WITH 2,4 DINITROPHENOL | | Descriptor: | 2,4-DINITROPHENOL, FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2002-02-22 | | Release date: | 2003-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Kinetic and Structural Basis of Reactivity of Pentaerythritol Tetranitrate Reductase with Nadph,2-Cyclohexenone Nitroesters and Nitroaromatic Explosives
J.Biol.Chem., 277, 2002
|
|
1GOL
 
 | |
1F2Z
 
 | |
1F35
 
 | |
1GVQ
 
 | |
1GWJ
 
 | | Morphinone reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, MORPHINONE REDUCTASE | | Authors: | Barna, T.M, Moody, P.C.E. | | Deposit date: | 2002-03-18 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Bacterial Morphinone Reductase and Properties of the C191A Mutant Enzyme.
J.Biol.Chem., 277, 2002
|
|
1H19
 
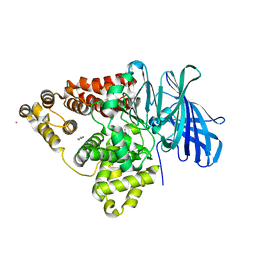 | | STRUCTURE OF [E271Q]LEUKOTRIENE A4 HYDROLASE | | Descriptor: | ACETIC ACID, IMIDAZOLE, LEUKOTRIENE A-4 HYDROLASE, ... | | Authors: | Rudberg, P.C, Tholander, F, Thunnissen, M.M.G.M, Haeggstrom, J.Z. | | Deposit date: | 2002-07-04 | | Release date: | 2002-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Leukotriene A4 Hydrolase/Aminopeptidase, Glutamate 271 is a Catalyticresidue with Specific Roles in Two Distinct Enzyme Mechanisms
J.Biol.Chem., 277, 2002
|
|
1H63
 
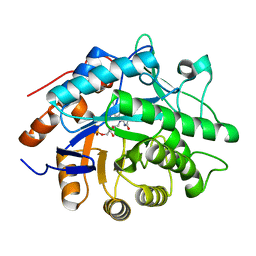 | | Structure of the reduced Pentaerythritol Tetranitrate Reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T.M, Moody, P.C.E. | | Deposit date: | 2001-06-04 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
1GNU
 
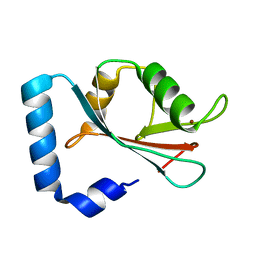 | | GABA(A) receptor associated protein GABARAP | | Descriptor: | GABARAP, NICKEL (II) ION | | Authors: | Knight, D, Harris, R, Moss, S, Driscoll, P.C, Keep, N.H. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-Ray Crystal Structure and Putative Ligand-Derived Peptide Binding Properties of Gamma-Aminobutyric Acid Receptor Type a Receptor-Associated Protein
J.Biol.Chem., 277, 2002
|
|
1H60
 
 | |
1H62
 
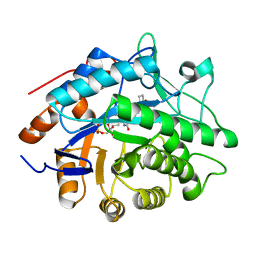 | | Structure of Pentaerythritol tetranitrate reductase in complex with 1,4-androstadien-3,17-dione | | Descriptor: | ANDROSTA-1,4-DIENE-3,17-DIONE, FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T.M, Moody, P.C.E. | | Deposit date: | 2001-06-04 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
7EBS
 
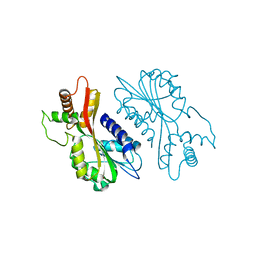 | | Crystal structure of juvenile hormone acid methyltransferase JHAMT from silkworm | | Descriptor: | Juvenile hormone acid methyltransferase | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, L, Xu, H.Y, Xia, Q.Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for juvenile hormone biosynthesis by the juvenile hormone acid methyltransferase.
J.Biol.Chem., 297, 2021
|
|
7EBX
 
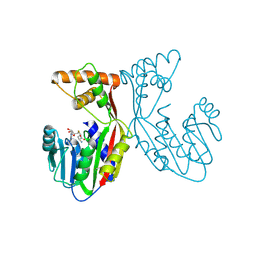 | | Crystal structure of juvenile hormone acid methyltransferase JHAMT in complex with S-adenosyl-L-homocysteine. | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, juvenile hormone acid methyltransferase | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, L, Xu, H.Y, Xia, Q.Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural basis for juvenile hormone biosynthesis by the juvenile hormone acid methyltransferase.
J.Biol.Chem., 297, 2021
|
|
7EC0
 
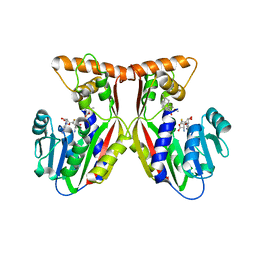 | | Crystal structure of juvenile hormone acid methyltransferase JHAMT in complex with S-Adenosyl homocysteine and methyl farnesoate | | Descriptor: | Juvenile hormone acid methyltransferase, Methyl farnesoate, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, L, Xu, H.Y, Xia, Q.Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Structural basis for juvenile hormone biosynthesis by the juvenile hormone acid methyltransferase.
J.Biol.Chem., 297, 2021
|
|
7EKE
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain F486L mutation complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKC
 
 | | Structure of SARS-CoV-2 Gamma variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
