1A0J
 
 | | CRYSTAL STRUCTURE OF A NON-PSYCHROPHILIC TRYPSIN FROM A COLD-ADAPTED FISH SPECIES. | | Descriptor: | BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Schroeder, H.-K, Willassen, N.P, Smalaas, A.O. | | Deposit date: | 1997-12-01 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a non-psychrophilic trypsin from a cold-adapted fish species.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
6MZZ
 
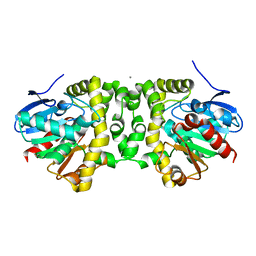 | | Fluoroacetate dehalogenase, room temperature structure, using first 1 degree of total 3 degree oscillation | | Descriptor: | CALCIUM ION, Fluoroacetate dehalogenase | | Authors: | Finke, A.D, Wierman, J.L, Pare-Labrosse, O, Sarrachini, A, Besaw, J, Mehrabi, P, Gruner, S.M, Miller, R.J.D. | | Deposit date: | 2018-11-06 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fixed-target serial oscillation crystallography at room temperature.
IUCrJ, 6, 2019
|
|
7PMM
 
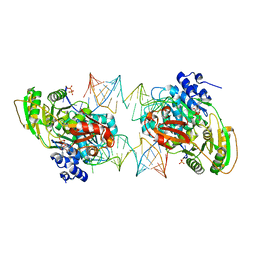 | |
7ZPP
 
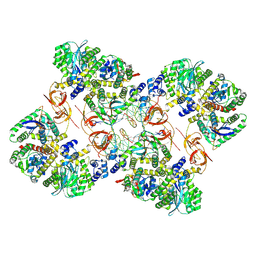 | | Cryo-EM structure of the MVV CSC intasome at 4.5A resolution | | Descriptor: | Integrase, vDNA, non-transferred strand, ... | | Authors: | Ballandras-Colas, A, Maskell, D, Pye, V.E, Locke, J, Swuec, S, Kotecha, A, Costa, A, Cherepanov, P. | | Deposit date: | 2022-04-28 | | Release date: | 2022-05-11 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | A supramolecular assembly mediates lentiviral DNA integration
Science, 355, 2017
|
|
6N3Z
 
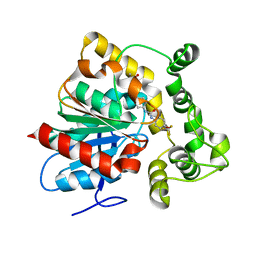 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 4 | | Descriptor: | Epoxide hydrolase TrEH, N-methyl-4-{[trans-4-({[4-(trifluoromethoxy)phenyl]carbamoyl}amino)cyclohexyl]oxy}benzamide | | Authors: | de Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.238 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
6N5H
 
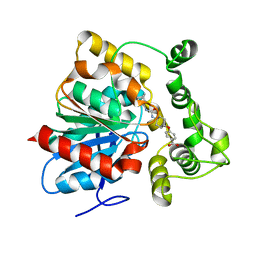 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 5 | | Descriptor: | 4-[(trans-4-{[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]dec-1-ylcarbamoyl]amino}cyclohexyl)oxy]benzoic acid, Epoxide hydrolase TrEH | | Authors: | Oliveira, G.S, Adriano, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors
Int. J. Biol. Macromol., 129, 2019
|
|
8CC2
 
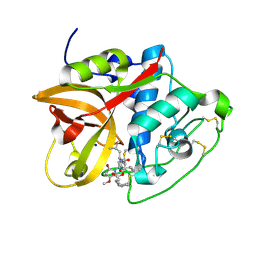 | | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide A | | Descriptor: | Cathepsin B-like peptidase (C01 family), SODIUM ION, gallinamide A, ... | | Authors: | Rubesova, P, Brynda, J, Mares, M, Gerwick, W.H. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Nature-Inspired Gallinamides Are Potent Antischistosomal Agents: Inhibition of the Cathepsin B1 Protease Target and Binding Mode Analysis.
Acs Infect Dis., 10, 2024
|
|
8CCU
 
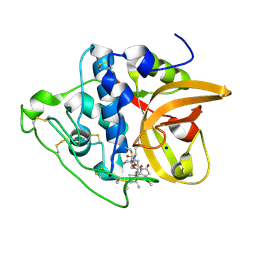 | | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide analog 1 | | Descriptor: | Cathepsin B-like peptidase (C01 family), SODIUM ION, [(2~{S})-1-[[(2~{S})-1-[[(2~{S})-5-[(2~{S})-3-methoxy-2-(2-methylpropyl)-5-oxidanylidene-2~{H}-pyrrol-1-yl]-5-oxidanylidene-pentan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl] (2~{S},3~{S})-2-(dimethylamino)-3-methyl-pentanoate | | Authors: | Rubesova, P, Brynda, J, Gerwick, W.H, Mares, M. | | Deposit date: | 2023-01-27 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nature-Inspired Gallinamides Are Potent Antischistosomal Agents: Inhibition of the Cathepsin B1 Protease Target and Binding Mode Analysis.
Acs Infect Dis., 10, 2024
|
|
3CAP
 
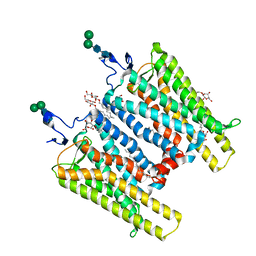 | | Crystal Structure of Native Opsin: the G Protein-Coupled Receptor Rhodopsin in its Ligand-free State | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Park, J.H, Scheerer, P, Hofmann, K.P, Choe, H.-W, Ernst, O.P. | | Deposit date: | 2008-02-20 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the ligand-free G-protein-coupled receptor opsin
Nature, 454, 2008
|
|
1AHS
 
 | |
4WB4
 
 | | wt SA11 NSP4_CCD | | Descriptor: | CALCIUM ION, Non-structural glycoprotein NSP4 | | Authors: | Viskovska, M, Sastri, N.P, Hyser, J.M, Tanner, M.R, Horton, L.B, Sankaran, B, Prasad, B.V.V, Estes, M.K. | | Deposit date: | 2014-09-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Plasticity of the Coiled-Coil Domain of Rotavirus NSP4.
J.Virol., 88, 2014
|
|
4R4D
 
 | | Racemic crystal structure of a magnesium-bound B-DNA duplex | | Descriptor: | 5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*G)-3', MAGNESIUM ION, SODIUM ION | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Huc, I. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Racemic DNA crystallography.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4WE9
 
 | | The crystal structure of hemagglutinin from influenza virus A/Victoria/361/2011 in complex with 3'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins.
Virology, 477C, 2015
|
|
4V2I
 
 | | Biochemical characterization and structural analysis of a new cold- active and salt tolerant esterase from the marine bacterium Thalassospira sp | | Descriptor: | ESTERASE/LIPASE, MAGNESIUM ION | | Authors: | Santi, C.D, Leiros, H.-K.S, Scala, A.D, Pascale, D.D, Altermark, B, Willassen, N.-P. | | Deposit date: | 2014-10-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | Biochemical Characterization and Structural Analysis of a New Cold-Active and Salt-Tolerant Esterase from the Marine Bacterium Thalassospira Sp.
Extremophiles, 20, 2016
|
|
7PFX
 
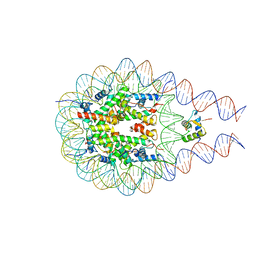 | |
7PEY
 
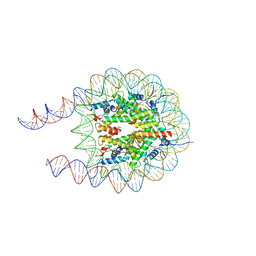 | |
4WE8
 
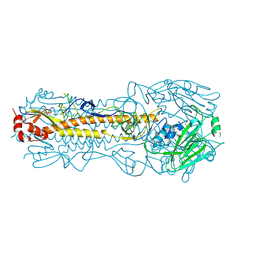 | | The crystal structure of hemagglutinin of influenza virus A/Victoria/361/2011 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins.
Virology, 477C, 2015
|
|
7PEZ
 
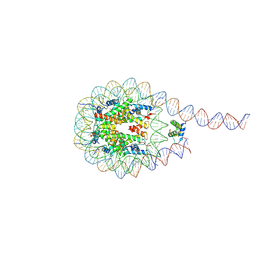 | |
7PF3
 
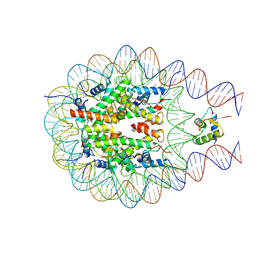 | |
4WDR
 
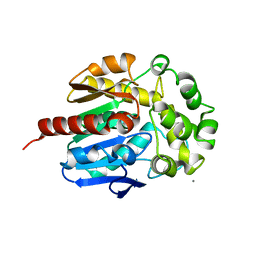 | | Crystal structure of haloalkane dehalogenase LinB 140A+143L+177W+211L mutant (LinB86) from Sphingobium japonicum UT26 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Degtjarik, O, Rezacova, P, Iermak, I, Chaloupkova, R, Damborsky, J, Kuta-Smatanova, I. | | Deposit date: | 2014-09-09 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of haloalkane dehalogenase LinB mutant (L177W) from Sphingobium japonicum UT26
Acs Catalysis, 2016
|
|
4WE5
 
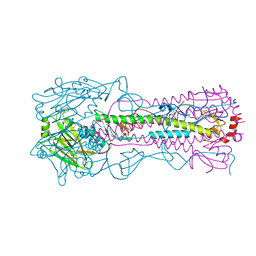 | | The crystal structure of hemagglutinin from A/Port Chalmers/1/1973 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins.
Virology, 477C, 2015
|
|
7PF4
 
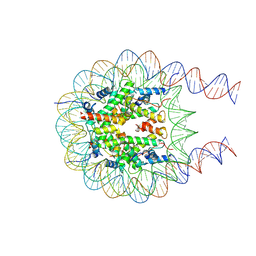 | |
4WEG
 
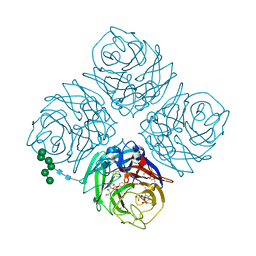 | | influenza virus neuraminidase N9 in complex 2,3-difluorosialic acid | | Descriptor: | (2R,3R,4R,5R,6R)-5-acetamido-2,3-difluoro-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]tetrahydro-2H-pyran-2-carboxylic acid, (3R,4R,5R,6R)-5-(acetylamino)-3-fluoro-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]-3,4,5,6-tetrahydropyranium-2-carboxylate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Streltsov, V.A, Pilling, P, Barrett, S, McKimm-Breschkin, J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-09-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalytic mechanism and novel receptor binding sites of human parainfluenza virus type 3 hemagglutinin-neuraminidase (hPIV3 HN)
Antiviral Res., 123, 2015
|
|
6N02
 
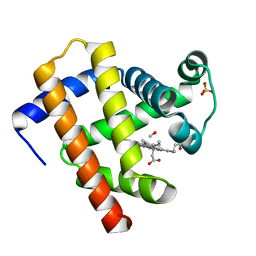 | | CO-bound Sperm Whale Myoglobin, room temperature structure, first 2 degrees of 5 degree total oscillation | | Descriptor: | CARBON MONOXIDE, Myoglobin, CO-bound, ... | | Authors: | Finke, A.D, Wierman, J.L, Pare-Labrosse, O, Sarrachini, A, Besaw, J, Mehrabi, P, Gruner, S.M, Miller, R.J.D. | | Deposit date: | 2018-11-06 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fixed-target serial oscillation crystallography at room temperature.
IUCrJ, 6, 2019
|
|
6Y2L
 
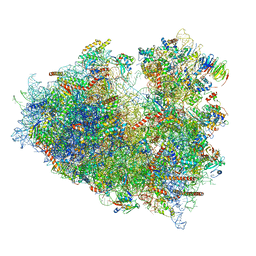 | | Structure of human ribosome in POST state | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Bhaskar, V, Schenk, A.D, Cavadini, S, von Loeffelholz, O, Natchiar, S.K, Klaholz, B.P, Chao, J.A. | | Deposit date: | 2020-02-16 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Dynamics of uS19 C-Terminal Tail during the Translation Elongation Cycle in Human Ribosomes.
Cell Rep, 31, 2020
|
|
