3KIA
 
 | | Crystal structure of mannosyl-3-phosphoglycerate synthase from Rubrobacter xylanophilus | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Macedo-Ribeiro, S, Pereira, P.J.B, Empadinhas, N, da Costa, M.S. | | Deposit date: | 2009-11-01 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functional and structural characterization of a novel mannosyl-3-phosphoglycerate synthase from Rubrobacter xylanophilus reveals its dual substrate specificity
Mol.Microbiol., 79, 2011
|
|
2OK5
 
 | | Human Complement factor B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement factor B, ... | | Authors: | Milder, F.J, Gomes, L, Schouten, A, Janssen, B.J.C, Huizinga, E.G, Romijn, R.A, Hemrika, W, Roos, A, Daha, M.R, Gros, P. | | Deposit date: | 2007-01-16 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Factor B structure provides insights into activation of the central protease of the complement system.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1Q25
 
 | | Crystal structure of N-terminal 3 domains of CI-MPR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Olson, L.J, Dahms, N.M, Kim, J.-J.P. | | Deposit date: | 2003-07-23 | | Release date: | 2004-06-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of uPAR, plasminogen, and sugar-binding sites of the 300 kDa mannose 6-phosphate receptor
Embo J., 23, 2004
|
|
2YIM
 
 | | The enolisation chemistry of a thioester-dependent racemase: the 1.4 A crystal structure of a complex with a planar reaction intermediate analogue | | Descriptor: | 2-METHYLACETOACETYL COA, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Sharma, S, Bhaumik, P, Venkatesan, R, Hiltunen, J.K, Conzelmann, E, Juffer, A.H, Wierenga, R.K. | | Deposit date: | 2011-05-16 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | The Enolization Chemistry of a Thioester-Dependent Racemase: The 1.4 A Crystal Structure of a Reaction Intermediate Complex Characterized by Detailed Qm/Mm Calculations.
J Phys Chem B, 116, 2012
|
|
4J6O
 
 | | Crystal Structure of the Phosphatase Domain of C. thermocellum (Bacterial) PnkP | | Descriptor: | CITRIC ACID, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Wang, L, Smith, P, Shuman, S. | | Deposit date: | 2013-02-11 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the 2',3' phosphatase component of the bacterial Pnkp-Hen1 RNA repair system.
Nucleic Acids Res., 41, 2013
|
|
2JT8
 
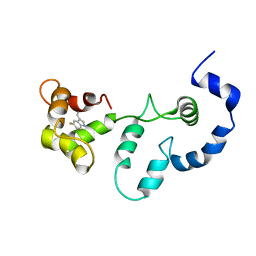 | | Solution structure of the F153-to-5-flurotryptophan mutant of human cardiac troponin C | | Descriptor: | Troponin C, slow skeletal and cardiac muscles | | Authors: | Wang, X, Mercier, P, Letourneau, P, Sykes, B.D. | | Deposit date: | 2007-07-20 | | Release date: | 2007-08-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies
Protein Sci., 14, 2005
|
|
3HGF
 
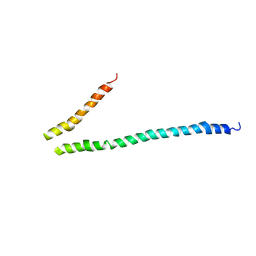 | | Expression, purification, spectroscopical and crystallographical studies of segments of the nucleotide binding domain of the reticulocyte binding protein Py235 of Plasmodium yoelii | | Descriptor: | Rhoptry protein fragment | | Authors: | Gruber, A, Manimekalai, M.S.S, Balakrishna, A.M, Hunke, C, Jeyakanthan, J, Preiser, P.R, Gruber, G. | | Deposit date: | 2009-05-13 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural determination of functional units of the nucleotide binding domain (NBD94) of the reticulocyte binding protein Py235 of Plasmodium yoelii
Plos One, 5, 2010
|
|
2JGY
 
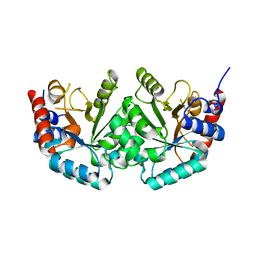 | | The crystal structure of human orotidine-5'-decarboxylase domain of human uridine monophosphate synthetase (UMPS) | | Descriptor: | UMP SYNTHASE | | Authors: | Moche, M, Ogg, D, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Johansson, I, Karlberg, T, Kosinska, U, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Uppenberg, J, Upsten, M, Thorsell, A.G, van den Berg, S, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-16 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of Human Orotidine-5'-Decarboxylase Domain of Human Uridine Monophosphate Synthetase (Umps)
To be Published
|
|
2YNC
 
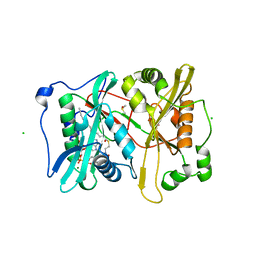 | | Plasmodium vivax N-myristoyltransferase in complex with YnC12-CoA thioester. | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | Authors: | Wright, M.H, Clough, B, Rackham, M.D, Brannigan, J.A, Grainger, M, Bottrill, A.R, Heal, W.P, Broncel, M, Serwa, R.A, Mann, D, Leatherbarrow, R.J, Wilkinson, A.J, Holder, A.A, Tate, E.W. | | Deposit date: | 2012-10-13 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Validation of N-Myristoyltransferase as an Antimalarial Drug Target Using an Integrated Chemical Biology Approach.
Nat.Chem., 6, 2014
|
|
4G4J
 
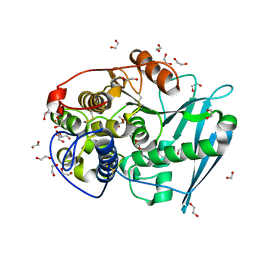 | | Crystal structure of glucuronoyl esterase S213A mutant from Sporotrichum thermophile in complex with methyl 4-O-methyl-beta-D-glucopyranuronate determined at 2.35 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4-O-methyl-glucuronoyl methylesterase, GLYCEROL, ... | | Authors: | Charavgi, M.D, Dimarogona, M, Topakas, E, Christakopoulos, P, Chrysina, E.D. | | Deposit date: | 2012-07-16 | | Release date: | 2013-01-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of a novel glucuronoyl esterase from Myceliophthora thermophila gives new insights into its role as a potential biocatalyst.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7MTT
 
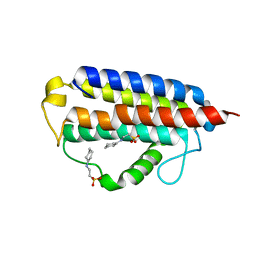 | |
4CYD
 
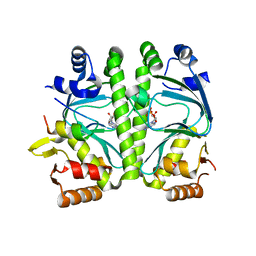 | | GlxR bound to cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, GLYCEROL, PROBABLE EXPRESSION TAG, ... | | Authors: | Townsend, P.D, Bott, M, Cann, M.J, Pohl, E. | | Deposit date: | 2014-04-10 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Crystal Structures of Apo and Camp-Bound Glxr from Corynebacterium Glutamicum Reveal Structural and Dynamic Changes Upon Camp Binding in Crp/Fnr Family Transcription Factors.
Plos One, 9, 2014
|
|
4N2G
 
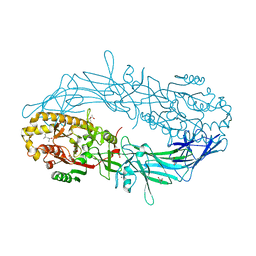 | | Crystal structure of Protein Arginine Deiminase 2 (D169A, 10 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4N28
 
 | | Crystal structure of Protein Arginine Deiminase 2 (1 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4RZ7
 
 | | Crystal Structure of PVX_084705 with bound PCI32765 | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, UNKNOWN ATOM OR ION, cGMP-dependent protein kinase, ... | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, He, H, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, El Bakkouri, M, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Crystal Structure of PVX_084705 with bound PCI32765
To be Published
|
|
2GJT
 
 | | Crystal structure of the human receptor phosphatase PTPRO | | Descriptor: | CHLORIDE ION, Receptor-type tyrosine-protein phosphatase PTPRO | | Authors: | Barr, A, Ugochukwu, E, Eswaran, J, Das, S, Niesen, F, Savitsky, P, Turnbull, A, Sundstrom, M, Arrowsmith, C, Edwards, A, Weigelt, J, von Delft, F, Papagrigoriou, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-31 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
5OWO
 
 | | Human cytoplasmic Dynein N-Terminus dimerization domain at 1.8 Angstrom resolution | | Descriptor: | CALCIUM ION, Cytoplasmic dynein 1 heavy chain 1, GLYCEROL, ... | | Authors: | Urnavicius, L, Lau, C.K, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
1DSA
 
 | | (+)-DUOCARMYCIN SA COVALENTLY LINKED TO DUPLEX DNA, NMR, 20 STRUCTURES | | Descriptor: | 4-HYDROXY-8-METHYL-6-(4,5,6-TRIMETHOXY-1H-INDOLE-2-CARBONYL)-3,6,7,8-TETRAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, DNA (5'-D(*GP*AP*CP*TP*AP*AP*TP*TP*GP*AP*C)-3', 5'-D(*GP*TP*CP*AP*AP*TP*TP*AP*GP*TP*C)-3') | | Authors: | Eis, P.S, Smith, J.A, Case, D.A, Chazin, W.J. | | Deposit date: | 1997-05-08 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution solution structure of a DNA duplex alkylated by the antitumor agent duocarmycin SA.
J.Mol.Biol., 272, 1997
|
|
1PLK
 
 | | CRYSTALLOGRAPHIC STUDIES ON P21H-RAS USING SYNCHROTRON LAUE METHOD: IMPROVEMENT OF CRYSTAL QUALITY AND MONITORING OF THE GTPASE REACTION AT DIFFERENT TIME POINTS | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Scheidig, A, Sanchez-Llorente, A, Lautwein, A, Pai, E.F, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Goody, R.S. | | Deposit date: | 1994-03-13 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic studies on p21(H-ras) using the synchrotron Laue method: improvement of crystal quality and monitoring of the GTPase reaction at different time points.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
2Q1D
 
 | | 2-keto-3-deoxy-D-arabinonate dehydratase complexed with magnesium and 2,5-dioxopentanoate | | Descriptor: | 2,5-DIOXOPENTANOIC ACID, 2-keto-3-deoxy-D-arabinonate dehydratase, MAGNESIUM ION | | Authors: | Barends, T, Brouns, S, Worm, P, Akerboom, J, Turnbull, A, Salmon, L. | | Deposit date: | 2007-05-24 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insight into substrate binding and catalysis of a novel 2-keto-3-deoxy-D-arabinonate dehydratase illustrates common mechanistic features of the FAH superfamily.
J.Mol.Biol., 379, 2008
|
|
1DYW
 
 | | Biochemical and structural characterization of a divergent loop cyclophilin from Caenorhabditis elegans | | Descriptor: | CYCLOPHILIN 3 | | Authors: | Dornan, J, Page, A.P, Taylor, P, Wu, S.Y, Winter, A.D, Husi, H, Walkinshaw, M.D. | | Deposit date: | 2000-02-10 | | Release date: | 2000-06-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and Structural Characterization of a Divergent Loop Cyclophilin from Caenorhabditis Elegans
J.Biol.Chem., 274, 1999
|
|
1RKY
 
 | | PPLO + Xe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Guss, J.M, Duff, A.P. | | Deposit date: | 2003-11-24 | | Release date: | 2004-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Using Xenon as a Probe for Dioxygen-binding Sites in Copper Amine Oxidases
J.Mol.Biol., 344, 2004
|
|
1DZ7
 
 | | Solution structure of the a-subunit of human chorionic gonadotropin [modeled without carbohydrate residues] | | Descriptor: | CHORIONIC GONADOTROPIN | | Authors: | Erbel, P.J.A, Karimi-Nejad, Y, De Beer, T, Boelens, R, Kamerling, J.P, Vliegenthart, J.F.G. | | Deposit date: | 2000-02-18 | | Release date: | 2000-02-29 | | Last modified: | 2019-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the alpha-subunit of human chorionic gonadotropin.
Eur.J.Biochem., 260, 1999
|
|
7VGJ
 
 | | Cryo-EM structure of the human P4-type flippase ATP8B1-CDC50A in the auto-inhibited E2Pi-PS state | | Descriptor: | Cell cycle control protein 50A, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, Phospholipid-transporting ATPase IC | | Authors: | Chen, M.T, Chen, Y, Chen, Z.P, Zhou, C.Z, Hou, W.T, Chen, Y. | | Deposit date: | 2021-09-16 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural insights into the activation of autoinhibited human lipid flippase ATP8B1 upon substrate binding.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4N2K
 
 | | Crystal structure of Protein Arginine Deiminase 2 (Q350A, 0 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-05 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
