5LYL
 
 | | Crystal structure of 1 in complex with tafCPB | | Descriptor: | (2~{S})-2-[[(2~{S})-1-(1-adamantylamino)-3-cyclohexyl-1-oxidanylidene-propan-2-yl]sulfamoylamino]-6-azanyl-hexanoic acid, Carboxypeptidase B, ZINC ION | | Authors: | Schreuder, H, Liesum, A, Loenze, P. | | Deposit date: | 2016-09-28 | | Release date: | 2016-10-26 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Sulfamide as Zinc Binding Motif in Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
J. Med. Chem., 59, 2016
|
|
3INW
 
 | | HSP90 N-TERMINAL DOMAIN with pochoxime A | | Descriptor: | (5E,9E,11E)-13-chloro-14,16-dihydroxy-3,4,7,8-tetrahydro-1H-2-benzoxacyclotetradecine-1,11(12H)-dione 11-[O-(2-oxo-2-piperidin-1-ylethyl)oxime], DIMETHYL SULFOXIDE, Heat shock protein HSP 90-alpha | | Authors: | Korndoerfer, I.P. | | Deposit date: | 2009-08-13 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Inhibition of HSP90 with pochoximes: SAR and structure-based insights.
Chembiochem, 10, 2009
|
|
1D7V
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 2,2-DIALKYLGLYCINE DECARBOXYLASE WITH NMA | | Descriptor: | N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-2-METHYLALANINE, POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
1CFF
 
 | | NMR SOLUTION STRUCTURE OF A COMPLEX OF CALMODULIN WITH A BINDING PEPTIDE OF THE CA2+-PUMP | | Descriptor: | CALCIUM ION, CALCIUM PUMP, CALMODULIN | | Authors: | Elshorst, B, Hennig, M, Foersterling, H, Diener, A, Maurer, M, Schulte, P, Schwalbe, H, Krebs, J, Schmid, H, Vorherr, T, Carafoli, E, Griesinger, C. | | Deposit date: | 1999-03-18 | | Release date: | 1999-09-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a complex of calmodulin with a binding peptide of the Ca2+ pump.
Biochemistry, 38, 1999
|
|
1CX9
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF BACTERIAL TRYPTOPHAN SYNTHASE WITH THE TRANSITION STATE ANALOGUE INHIBITOR 4-(2-AMINOPHENYLTHIO)-BUTYLPHOSPHONIC ACID | | Descriptor: | 4-(2-AMINOPHENYLTHIO)-BUTYLPHOSPHONIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Sachpatzidis, A, Dealwis, C, Lubetsky, J.B, Liang, P.H, Anderson, K.S, Lolis, E. | | Deposit date: | 1999-08-29 | | Release date: | 1999-12-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic studies of phosphonate-based alpha-reaction transition-state analogues complexed to tryptophan synthase.
Biochemistry, 38, 1999
|
|
6OLM
 
 | |
1CZ2
 
 | | SOLUTION STRUCTURE OF WHEAT NS-LTP COMPLEXED WITH PROSTAGLANDIN B2. | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN, PROSTAGLANDIN B2 | | Authors: | Tassin-Moindrot, S, Caille, A, Douliez, J.-P, Marion, D, Vovelle, F. | | Deposit date: | 1999-09-01 | | Release date: | 2000-09-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The wide binding properties of a wheat nonspecific lipid transfer protein. Solution structure of a complex with prostaglandin B2.
Eur.J.Biochem., 267, 2000
|
|
1D7R
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 2,2-DIALKYLGLYCINE DECARBOXYLASE WITH 5PA | | Descriptor: | N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID, POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
1CEK
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE MEMBRANE-EMBEDDED M2 CHANNEL-LINING SEGMENT FROM THE NICOTINIC ACETYLCHOLINE RECEPTOR BY SOLID-STATE NMR SPECTROSCOPY | | Descriptor: | PROTEIN (ACETYLCHOLINE RECEPTOR M2) | | Authors: | Marassi, F.M, Gesell, J.J, Kim, Y, Valente, A.P, Oblatt-Montal, M, Montal, M, Opella, S.J. | | Deposit date: | 1999-03-09 | | Release date: | 1999-03-11 | | Last modified: | 2023-12-27 | | Method: | SOLID-STATE NMR | | Cite: | Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy.
Nat.Struct.Biol., 6, 1999
|
|
7OOO
 
 | | The crystal structure of a DNA:RNA hybrid duplex sequence CTTTTCTTTG containing an LNA-Amide-LNA modification | | Descriptor: | DNA (5'-D(*CP*TP*(05A)P*TP*CP*TP*TP*TP*G)-3'), MAGNESIUM ION, RNA (5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3') | | Authors: | Thorpe, C, Hardwick, J, McDonough, M.A, Hall, J.P, Baker, Y.R, El-Sagheer, A.H, Brown, T. | | Deposit date: | 2021-05-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | An LNA-amide modification that enhances the cell uptake and activity of phosphorothioate exon-skipping oligonucleotides.
Nat Commun, 13, 2022
|
|
7OOS
 
 | | The crystal structure of a DNA:RNA hybrid duplex sequence CTTTTCTTTG | | Descriptor: | DNA (5'-D(*CP*TP*(05K)P*TP*CP*TP*TP*TP*G)-3'), RNA (5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3'), STRONTIUM ION | | Authors: | Thorpe, C, Hardwick, J, McDonough, M.A, Hall, J.P, Baker, Y.R, El-Sagheer, A.H, Brown, T. | | Deposit date: | 2021-05-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An LNA-amide modification that enhances the cell uptake and activity of phosphorothioate exon-skipping oligonucleotides.
Nat Commun, 13, 2022
|
|
7OZZ
 
 | | The crystal structure of a DNA:RNA hybrid duplex sequence CTTTTCTTTG with LNA-amide modification | | Descriptor: | DNA (5'-D(*CP*TP*(05H)P*TP*CP*TP*TP*TP*G)-3'), POTASSIUM ION, RNA (5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3') | | Authors: | Thorpe, C, Hardwick, J, McDonough, M.A, Hall, J.P, Baker, Y.R, El-Sagheer, A.H, Brown, T. | | Deposit date: | 2021-06-29 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An LNA-amide modification that enhances the cell uptake and activity of phosphorothioate exon-skipping oligonucleotides.
Nat Commun, 13, 2022
|
|
6OS1
 
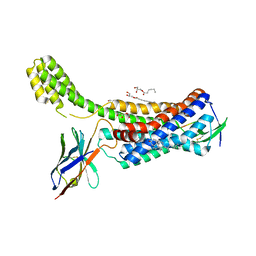 | | Structure of synthetic nanobody-stabilized angiotensin II type 1 receptor bound to TRV023 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Nanobody Nb.AT110i1_le, ... | | Authors: | Wingler, L.M, Staus, D.P, Skiba, M.A, McMahon, C, Kleinhenz, A.L.W, Lefkowitz, R.J, Kruse, A.C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR.
Science, 367, 2020
|
|
1D7U
 
 | | Crystal structure of the complex of 2,2-dialkylglycine decarboxylase with LCS | | Descriptor: | POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), SODIUM ION, ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
5LYF
 
 | | Crystal structure of 1 in complex with tafCPB | | Descriptor: | (2~{S})-6-azanyl-2-[[(2~{R})-1-[[(1~{R},2~{S},4~{S})-2-bicyclo[2.2.1]heptanyl]amino]-3-cyclohexyl-1-oxidanylidene-propan-2-yl]carbamoylamino]hexanoic acid, Carboxypeptidase B, ZINC ION | | Authors: | Schreuder, H, Liesum, A, Loenze, P. | | Deposit date: | 2016-09-28 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Sulfamide as Zinc Binding Motif in Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
J. Med. Chem., 59, 2016
|
|
1CE2
 
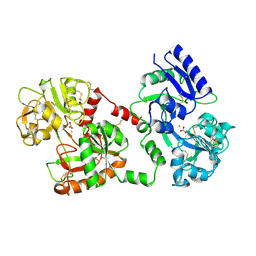 | | STRUCTURE OF DIFERRIC BUFFALO LACTOFERRIN AT 2.5A RESOLUTION | | Descriptor: | CARBONATE ION, FE (III) ION, PROTEIN (LACTOFERRIN) | | Authors: | Karthikeyan, S, Paramasivam, M, Yadav, S, Srinivasan, A, Singh, T.P. | | Deposit date: | 1999-03-13 | | Release date: | 1999-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of buffalo lactoferrin at 2.5 A resolution using crystals grown at 303 K shows different orientations of the N and C lobes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1D8A
 
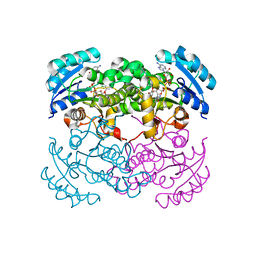 | | E. COLI ENOYL REDUCTASE/NAD+/TRICLOSAN COMPLEX | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Levy, C.W, Roujeinikova, A, Sedelnikova, S, Baker, P.J, Stuitje, A.R, Slabas, A.R, Rice, D.W, Rafferty, J.B. | | Deposit date: | 1999-10-21 | | Release date: | 1999-10-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of triclosan activity.
Nature, 398, 1999
|
|
3INX
 
 | | HSP90 N-TERMINAL DOMAIN with pochoxime B | | Descriptor: | (5E,9E,11E)-14,16-dihydroxy-3,4,7,8-tetrahydro-1H-2-benzoxacyclotetradecine-1,11(12H)-dione 11-[O-(2-oxo-2-piperidin-1-ylethyl)oxime], DIMETHYL SULFOXIDE, Heat shock protein HSP 90-alpha | | Authors: | Korndoerfer, I.P. | | Deposit date: | 2009-08-13 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inhibition of HSP90 with pochoximes: SAR and structure-based insights.
Chembiochem, 10, 2009
|
|
1D9E
 
 | | STRUCTURE OF E. COLI KDO8P SYNTHASE | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE SYNTHASE, SULFATE ION | | Authors: | Radaev, S, Dastidar, P, Patel, M, Woodard, R.W, Gatti, D.L. | | Deposit date: | 1999-10-27 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of 3-deoxy-D-manno-octulosonate 8-phosphate synthase.
J.Biol.Chem., 275, 2000
|
|
6OLL
 
 | |
7OGV
 
 | | A self-complementary DNA dodecamer duplex contaning 5-hydroxymethylcitosine | | Descriptor: | DNA (5'-D(*(P*GP*CP*GP*TP*(DH)P*GP*AP*CP*GP*CP*G-3') | | Authors: | Battistini, F, Dans, P.D, Terrazas, M, Castellazzi, C.L, Portella, G, Labrador, M, Villegas, N, Brun-Heath, I, Gonzalez, C, Orozco, M. | | Deposit date: | 2021-05-07 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Impact of the HydroxyMethylCytosine epigenetic signature on DNA structure and function.
Plos Comput.Biol., 17, 2021
|
|
8CO8
 
 | | Structure of West Nile Virus NS2B-NS3 protease | | Descriptor: | Serine protease subunit NS2B,Serine protease/Helicase NS3 | | Authors: | Fairhead, M, Godoy, A.S, Koekemoer, L, Balcomb, B.H, Lithgo, R.M, Aschenbrenner, J.C, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-02-27 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of West Nile Virus NS2B-NS3 protease - to be published
To Be Published
|
|
7PIL
 
 | | Cryo-EM structure of the Rhodobacter sphaeroides RC-LH1-PufXY monomer complex at 2.5 A | | Descriptor: | (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, BACTERIOCHLOROPHYLL A, ... | | Authors: | Qian, P, Hunter, C.N. | | Deposit date: | 2021-08-20 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of the monomeric Rhodobacter sphaeroides RC-LH1 core complex at 2.5 angstrom.
Biochem.J., 478, 2021
|
|
8JEY
 
 | |
7PMK
 
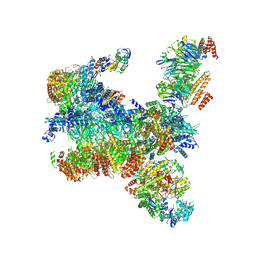 | | S. cerevisiae replisome-SCF(Dia2) complex bound to double-stranded DNA (conformation I) | | Descriptor: | Cell division control protein 45,Cell division control protein 45, Chromosome segregation in meiosis protein 3, DNA helicase, ... | | Authors: | Jenkyn-Bedford, M, Yeeles, J.T.P, Deegan, T.D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A conserved mechanism for regulating replisome disassembly in eukaryotes.
Nature, 600, 2021
|
|
