6FJP
 
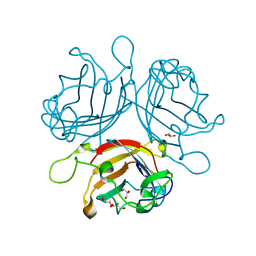 | | Adenovirus species 26 knob protein, high resolution, High pH | | Descriptor: | 1,2-ETHANEDIOL, Fiber, GLYCEROL | | Authors: | Rizkallah, P.J, Parker, A.L, Baker, A.T. | | Deposit date: | 2018-01-22 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Diversity within the adenovirus fiber knob hypervariable loops influences primary receptor interactions.
Biorxiv, 2019
|
|
5XTN
 
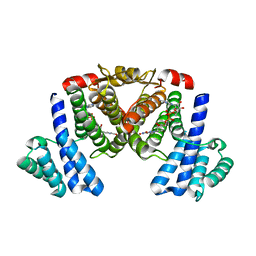 | |
5E1X
 
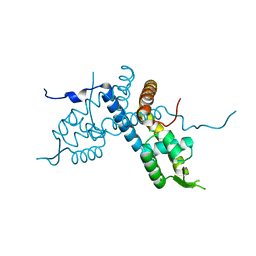 | | Crystal structure of the organohalide sensing RdhR-CbdbA1625 transcriptional regulator in the 3,4-dichlorophenol bound form | | Descriptor: | 3,4-dichlorophenol, Transcriptional regulator, MarR family | | Authors: | Quezada, C.P, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2015-09-30 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal structures of RdhRCbdbA1625 provide insight into sensing of chloroaromatic compounds by MarR-type regulators
TO BE PUBLISHED
|
|
4K38
 
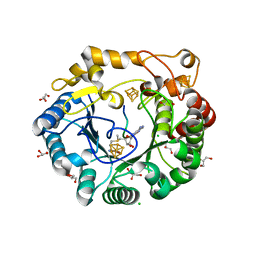 | | Native anSMEcpe with bound AdoMet and Kp18Cys peptide | | Descriptor: | Anaerobic sulfatase-maturating enzyme, CHLORIDE ION, GLYCEROL, ... | | Authors: | Goldman, P.J, Drennan, C.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.831 Å) | | Cite: | X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5S81
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010947a | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type-1, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S7R
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010918a | | Descriptor: | 1,2-ETHANEDIOL, 1lambda~6~,2-thiazetidine-1,1-dione, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | XChem group deposition
To Be Published
|
|
6JDI
 
 | | Central domain of FleQ H287N mutant in complex with ATPgS and Mg | | Descriptor: | MAGNESIUM ION, Nitrogen assimilation regulatory protein, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Jain, D, Banerjee, P, Chanchal | | Deposit date: | 2019-02-01 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Sensor I Regulated ATPase Activity of FleQ Is Essential for Motility to Biofilm Transition inPseudomonas aeruginosa.
Acs Chem.Biol., 14, 2019
|
|
5S86
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010952a | | Descriptor: | 1,2-ETHANEDIOL, 1-aminocyclopropane-1-carboxamide, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S80
 
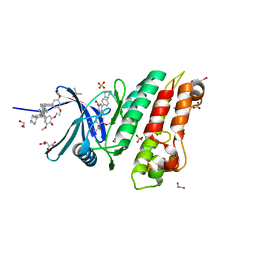 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010946a | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type-1, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5X4X
 
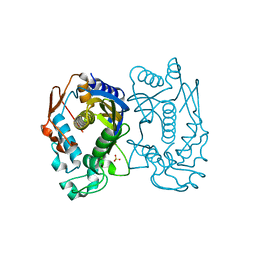 | | Mutant human thymidylate synthase A191K crystallized in a sulfate-containing condition | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Chen, D, Jansson, A, Larsson, A, Nordlund, P. | | Deposit date: | 2017-02-14 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analyses of human thymidylate synthase reveal a site that may control conformational switching between active and inactive states
J. Biol. Chem., 292, 2017
|
|
6JDL
 
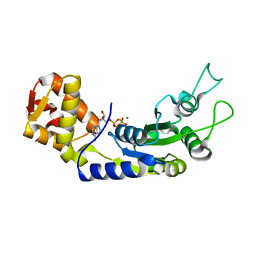 | | Central domain of FleQ H287A mutant in complex with ATPgS and Mg | | Descriptor: | MAGNESIUM ION, Nitrogen assimilation regulatory protein, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Jain, D, Banerjee, P, Chanchal | | Deposit date: | 2019-02-01 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Sensor I Regulated ATPase Activity of FleQ Is Essential for Motility to Biofilm Transition inPseudomonas aeruginosa.
Acs Chem.Biol., 14, 2019
|
|
5S7J
 
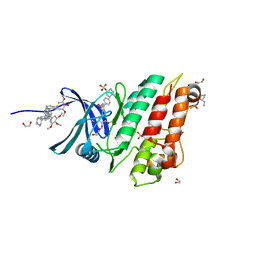 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM000893d | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2R)-oxolan-2-yl]methanamine, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S77
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with XS035133b | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type-1, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S7U
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010938a | | Descriptor: | (4S)-1-methylimidazolidin-4-amine, 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | XChem group deposition
To Be Published
|
|
3M5N
 
 | |
5S7B
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM000329d | | Descriptor: | (3R)-thiolane-3-carboxylic acid, 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S82
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with XS035128c | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type-1, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S7E
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010930a | | Descriptor: | (3R)-1,2-oxazolidine-3-carbonitrile, 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5XWJ
 
 | | Crystal Structure of Porcine pancreatic trypsin with tripeptide inhibitor, TRE, at pH 7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Acetylated-THR-ARG-GLU Inhibitor, CALCIUM ION, ... | | Authors: | Saikhedkar, N.S, Bhoite, A.S, Giri, A.P, Kulkarni, K.A. | | Deposit date: | 2017-06-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tripeptides derived from reactive centre loop of potato type II protease inhibitors preferentially inhibit midgut proteases of Helicoverpa armigera.
Insect Biochem. Mol. Biol., 95, 2018
|
|
6FGN
 
 | | Solution Structure of p300Taz2-p63TA | | Descriptor: | Histone acetyltransferase p300,Tumor protein 63, ZINC ION | | Authors: | Gebel, J, Kazemi, S, Lohr, F, Guntert, P, Dotsch, V. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Regulation of the Activity in the p53 Family Depends on the Organization of the Transactivation Domain.
Structure, 26, 2018
|
|
3ZJM
 
 | | Ile(149)G11Phe mutation of M.acetivorans protoglobin in complex with cyanide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CYANIDE ION, GLYCEROL, ... | | Authors: | Pesce, A, Tilleman, L, Donne, J, Aste, E, Ascenzi, P, Ciaccio, C, Coletta, M, Moens, L, Viappiani, C, Dewilde, S, Bolognesi, M, Nardini, M. | | Deposit date: | 2013-01-18 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Haem-Distal Site Plasticity in Methanosarcina Acetivorans Protoglobin.
Plos One, 8, 2013
|
|
2JDQ
 
 | | C-terminal domain of influenza A virus polymerase PB2 subunit in complex with human importin alpha5 | | Descriptor: | IMPORTIN ALPHA-1 SUBUNIT, POLYMERASE BASIC PROTEIN 2 | | Authors: | Tarendeau, F, Guilligay, D, Mas, P, Boulo, S, Baudin, F, Ruigrok, R.W.H, Hart, D.J, Cusack, S. | | Deposit date: | 2007-01-11 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Nuclear Import Function of the C- Terminal Domain of Influenza Virus Polymerase Pb2 Subunit
Nat.Struct.Mol.Biol., 14, 2007
|
|
3ZSQ
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | 5-[[[2-[[(1S)-1-(4-METHOXYPHENYL)BUTYL]CARBAMOYL]PHENYL]METHYL-PROP-2-ENYL-AMINO]METHYL]-1,3-BENZODIOXOLE-4-CARBOXYLIC ACID, ACETIC ACID, GLYCEROL, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-06-30 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
6JHG
 
 | | Crystal structure of apo Pullulanase from Paenibacillus barengoltzii in space group P212121 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pulullanase | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Crystal structure of apo Pullulanase from Paenibacillus barengoltzii in space group P212121
To Be Published
|
|
5XBM
 
 | | Structure of SCARB2-JL2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome membrane protein 2, ... | | Authors: | Zhang, X, Yang, P, Wang, N, Zhang, J, Li, J, Guo, H, Yin, X, Rao, Z, Wang, X, Zhang, L. | | Deposit date: | 2017-03-20 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | The binding of a monoclonal antibody to the apical region of SCARB2 blocks EV71 infection.
Protein Cell, 8, 2017
|
|
