7BLR
 
 | | Vps35/Vps29 arch of fungal membrane-assembled retromer:Vps5 (SNX-BAR) complex. | | Descriptor: | Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Leneva, N, Kovtun, O, Morado, D.R, Briggs, J.A.G, Owen, D.J. | | Deposit date: | 2021-01-18 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Architecture and mechanism of metazoan retromer:SNX3 tubular coat assembly.
Sci Adv, 7, 2021
|
|
7BLP
 
 | | Vps35/Vps29 arch of fungal membrane-assembled retromer:Grd19 complex | | Descriptor: | Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Leneva, N, Kovtun, O, Morado, D.R, Briggs, J.A.G, Owen, D.J. | | Deposit date: | 2021-01-18 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Architecture and mechanism of metazoan retromer:SNX3 tubular coat assembly.
Sci Adv, 7, 2021
|
|
7BLN
 
 | | VPS35/VPS29 arch of metazoan membrane-assembled retromer:SNX3 complex modelled with human proteins | | Descriptor: | Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Leneva, N, Kovtun, O, Morado, D.R, Briggs, J.A.G, Owen, D.J. | | Deposit date: | 2021-01-18 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Architecture and mechanism of metazoan retromer:SNX3 tubular coat assembly.
Sci Adv, 7, 2021
|
|
7BLO
 
 | | VPS26 dimer region of metazoan membrane-assembled retromer:SNX3 complex modelled with human proteins | | Descriptor: | 2-(BUTANOYLOXY)-1-{[(HYDROXY{[2,3,4,6-TETRAHYDROXY-5-(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]METHYL}ETHYL BUTANOATE, C-term (residues 493-54) of Wls (fitted sequence corresponds to hDMT1-II), Sorting nexin-3, ... | | Authors: | Leneva, N, Kovtun, O, Morado, D.R, Briggs, J.A.G, Owen, D.J. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-03 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Architecture and mechanism of metazoan retromer:SNX3 tubular coat assembly.
Sci Adv, 7, 2021
|
|
7BLQ
 
 | | Vps26 dimer region of the fungal membrane-assembled retromer:Grd19 complex. | | Descriptor: | 2-(BUTANOYLOXY)-1-{[(HYDROXY{[2,3,4,6-TETRAHYDROXY-5-(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]METHYL}ETHYL BUTANOATE, Sorting nexin-3, The C-terminal portion of Kex2 cargo, ... | | Authors: | Leneva, N, Kovtun, O, Morado, D.R, Briggs, J.A.G, Owen, D.J. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | Architecture and mechanism of metazoan retromer:SNX3 tubular coat assembly.
Sci Adv, 7, 2021
|
|
2BP5
 
 | | MU2 ADAPTIN SUBUNIT (AP50) OF AP2 ADAPTOR (SECOND DOMAIN), COMPLEXED WITH NON-CANONICAL INTERNALIZATION PEPTIDE VEDYEQGLSG | | Descriptor: | CLATHRIN COAT ASSEMBLY PROTEIN AP50, P2X PURINOCEPTOR 4 | | Authors: | Royle, S.J, Evans, P.R, Owen, D.J, Murrell-Lagnado, R.D. | | Deposit date: | 2005-04-18 | | Release date: | 2005-07-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Non-Canonical Yxxg-Phi Endocytic Motifs: Recognition by Ap2 and Preferential Utilization in P2X4 Receptors.
J.Cell.Sci, 118, 2005
|
|
1QL6
 
 | | THE CATALYTIC MECHANISM OF PHOSPHORYLASE KINASE PROBED BY MUTATIONAL STUDIES | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PHOSPHORYLASE KINASE, ... | | Authors: | Skamnaki, V.T, Owen, D.J, Noble, M.E.M, Lowe, E.D, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1999-08-24 | | Release date: | 1999-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic Mechanism of Phosphorylase Kinase Probed by Mutational Studies.
Biochemistry, 38, 1999
|
|
5JP2
 
 | |
1R4X
 
 | | Crystal Structure Analys of the Gamma-COPI Appendage domain | | Descriptor: | Coatomer gamma subunit, MAGNESIUM ION | | Authors: | Watson, P.J, Frigerio, G, Collins, B.M, Duden, R, Owen, D.J. | | Deposit date: | 2003-10-09 | | Release date: | 2003-10-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gamma-COP appendage domain - structure and function
Traffic, 5, 2004
|
|
1UTC
 
 | | Clathrin terminal domain complexed with TLPWDLWTT | | Descriptor: | AMPHIPHYSIN, CLATHRIN HEAVY CHAIN | | Authors: | Miele, A.E, Evans, P.R, Owen, D.J. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two distinct interaction motifs in amphiphysin bind two independent sites on the clathrin terminal domain beta-propeller.
Nat. Struct. Mol. Biol., 11, 2004
|
|
1Z2W
 
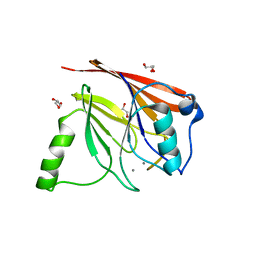 | | Crystal structure of mouse Vps29 complexed with Mn2+ | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Vacuolar protein sorting 29 | | Authors: | Collins, B.M, Skinner, C.F, Watson, P.J, Seaman, M.N.J, Owen, D.J. | | Deposit date: | 2005-03-10 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly
NAT.STRUCT.MOL.BIOL., 12, 2005
|
|
1Z2X
 
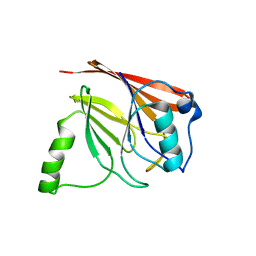 | | Crystal structure of mouse Vps29 | | Descriptor: | Vacuolar protein sorting 29 | | Authors: | Collins, B.M, Skinner, C.F, Watson, P.J, Seaman, M.N.J, Owen, D.J. | | Deposit date: | 2005-03-10 | | Release date: | 2005-06-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly
NAT.STRUCT.MOL.BIOL., 12, 2005
|
|
3ZYK
 
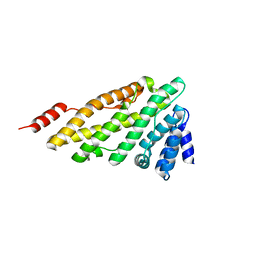 | | Structure of CALM (PICALM) ANTH domain | | Descriptor: | PHOSPHATIDYLINOSITOL-BINDING CLATHRIN ASSEMBLY PROTEIN | | Authors: | Miller, S.E, Sahlender, D.A, Graham, S.C, Honing, S, Robinson, M.S, Peden, A.A, Owen, D.J. | | Deposit date: | 2011-08-23 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM.
Cell, 147, 2011
|
|
3ZYM
 
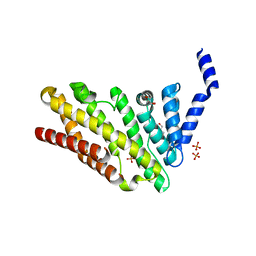 | | Structure of CALM (PICALM) in complex with VAMP8 | | Descriptor: | GLYCEROL, PHOSPHATE ION, PHOSPHATIDYLINOSITOL-BINDING CLATHRIN ASSEMBLY PROTEIN, ... | | Authors: | Miller, S.E, Sahlender, D.A, Graham, S.C, Honing, S, Robinson, M.S, Peden, A.A, Owen, D.J. | | Deposit date: | 2011-08-23 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM.
Cell, 147, 2011
|
|
3ZYL
 
 | | Structure of a truncated CALM (PICALM) ANTH domain | | Descriptor: | PHOSPHATIDYLINOSITOL-BINDING CLATHRIN ASSEMBLY PROTEIN | | Authors: | Miller, S.E, Sahlender, D.A, Graham, S.C, Honing, S, Robinson, M.S, Peden, A.A, Owen, D.J. | | Deposit date: | 2011-08-23 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM.
Cell, 147, 2011
|
|
3I2W
 
 | |
4AFI
 
 | | Complex between Vamp7 longin domain and fragment of delta-adaptin from AP3 | | Descriptor: | AP-3 COMPLEX SUBUNIT DELTA-1, VESICLE-ASSOCIATED MEMBRANE PROTEIN 7, PRASEODYMIUM ION | | Authors: | Kent, H.M, Evans, P.R, Luzio, J.P, Peden, A.A, Owen, D.J. | | Deposit date: | 2012-01-19 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of the Intracellular Sorting of the Snare Vamp7 by the Ap3 Adaptor Complex
Dev.Cell, 22, 2012
|
|
2YNP
 
 | | yeast betaprime COP 1-604 with KTKTN motif | | Descriptor: | COATOMER SUBUNIT BETA', KTKTN MOTIF | | Authors: | Jackson, L.P, Lewis, M, Kent, H.M, Edeling, M.A, Evans, P.R, Duden, R, Owen, D.J. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.962 Å) | | Cite: | Molecular Basis for Recognition of Dilysine Trafficking Motifs by Copi.
Dev.Cell, 23, 2012
|
|
2YNN
 
 | | yeast betaprime COP 1-304 with KTKTN motif | | Descriptor: | COATOMER SUBUNIT BETA', KTKTN MOTIF, SULFATE ION | | Authors: | Jackson, L.P, Lewis, M, Kent, H.M, Edeling, M.A, Evans, P.R, Duden, R, Owen, D.J. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Molecular Basis for Recognition of Dilysine Trafficking Motifs by Copi.
Dev.Cell, 23, 2012
|
|
2YNO
 
 | | yeast betaprime COP 1-304H6 | | Descriptor: | COATOMER SUBUNIT BETA', POLY ALA | | Authors: | Jackson, L.P, Lewis, M, Kent, H.M, Edeling, M.A, Evans, P.R, Duden, R, Owen, D.J. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Recognition of Dilysine Trafficking Motifs by Copi.
Dev.Cell, 23, 2012
|
|
1OM9
 
 | | Structure of the GGA1-appendage in complex with the p56 binding peptide | | Descriptor: | 15-mer peptide fragment of p56, ADP-ribosylation factor binding protein GGA1 | | Authors: | Collins, B.M, Praefcke, G.J.K, Robinson, M.S, Owen, D.J. | | Deposit date: | 2003-02-25 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for binding of accessory proteins by the appendage domain of GGAs
Nat.Struct.Biol., 10, 2003
|
|
1NA8
 
 | | Crystal structure of ADP-ribosylation factor binding protein GGA1 | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Lui, W.W, Collins, B.M, Hirst, J, Motley, A, Millar, C, Schu, P, Owen, D.J, Robinson, M.S. | | Deposit date: | 2002-11-27 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding partners for the COOH-terminal appendage domains of the GGAs and gamma-adaptin
Mol.Cell.Biol., 14, 2003
|
|
6QH5
 
 | | AP2 clathrin adaptor mu2T156-phosphorylated core in closed conformation | | Descriptor: | AP-2 complex subunit alpha, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Wrobel, A.G, Owen, D.J, McCoy, A.J, Evans, P.R. | | Deposit date: | 2019-01-15 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
6QH6
 
 | | AP2 clathrin adaptor core with two cargo peptides in open+ conformation | | Descriptor: | AP-2 complex subunit alpha, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Wrobel, A.G, Owen, D.J, McCoy, A.J, Evans, P.R. | | Deposit date: | 2019-01-15 | | Release date: | 2019-09-04 | | Last modified: | 2022-12-07 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
6QH7
 
 | | AP2 clathrin adaptor mu2T156-phosphorylated core with two cargo peptides in open+ conformation | | Descriptor: | ADAPTOR-RELATED PROTEIN COMPLEX 2, MU 2 SUBUNIT, C-TERMINAL DOMAIN, ... | | Authors: | Wrobel, A.G, Owen, D.J, McCoy, A.J, Evans, P.R. | | Deposit date: | 2019-01-15 | | Release date: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
