1K76
 
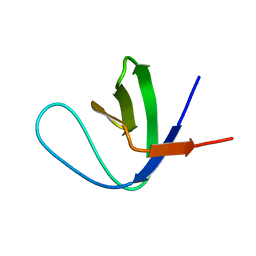 | | Solution Structure of the C-terminal Sem-5 SH3 Domain (Minimized Average Structure) | | Descriptor: | SEX MUSCLE ABNORMAL PROTEIN 5 | | Authors: | Ferreon, J, Volk, D, Luxon, B, Gorenstein, D, Hilser, V. | | Deposit date: | 2001-10-18 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Dynamics and Thermodynamics of the Native State Ensemble
of Sem-5 C-Terminal SH3 Domain
Biochemistry, 42, 2003
|
|
1K8L
 
 | | XBY6: An analog of CK14 containing 6 dithiophosphate groups | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1K8J
 
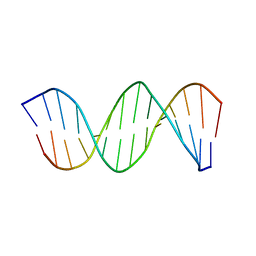 | | NMR STRUCTURE OF THE CK14 DNA DUPLEX: A PORTION OF THE KNOWN NF-kB SEQUENCE CK1 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1K8N
 
 | | NMR structure of the XBY2 DNA duplex, an analog of CK14 containing phosphorodithioate groups at C22 and C24 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1EKW
 
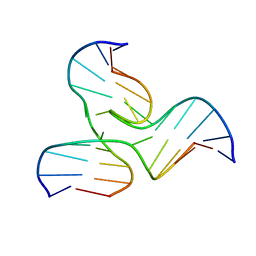 | | NMR STRUCTURE OF A DNA THREE-WAY JUNCTION | | Descriptor: | DNA (5'-D(*CP*GP*GP*TP*GP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*CP*AP*CP*CP*G)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*TP*CP*GP*CP*AP*GP*C)-3') | | Authors: | Thiviyanathan, V, Luxon, B.A, Leontis, N.B, Donne, D, Gorenstein, D.G. | | Deposit date: | 2000-03-09 | | Release date: | 2000-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hybrid-hybrid matrix structural refinement of a DNA three-way junction from 3D NOESY-NOESY.
J.Biomol.NMR, 14, 1999
|
|
1Z3R
 
 | | Solution structure of the Omsk Hemhorraghic Fever Envelope Protein Domain III | | Descriptor: | polyprotein | | Authors: | Volk, D.E, Chavez, L, Beasley, D.W, Barrett, A.D, Gorenstein, D.G. | | Deposit date: | 2005-03-14 | | Release date: | 2006-03-14 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the envelope protein domain III of Omsk hemorrhagic fever virus.
Virology, 351, 2006
|
|
1KFZ
 
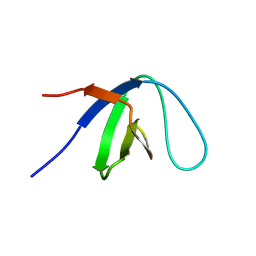 | | Solution Structure of C-terminal Sem-5 SH3 Domain (Ensemble of 16 Structures) | | Descriptor: | SEX MUSCLE ABNORMAL PROTEIN 5 | | Authors: | Ferreon, J.C, Volk, D.E, Luxon, B.A, Gorenstein, D, Hilser, V.J. | | Deposit date: | 2001-11-24 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Dynamics and Thermodynamics of the Native State Ensemble
of Sem-5 C-Terminal SH3 Domain
Biochemistry, 42, 2003
|
|
1N2W
 
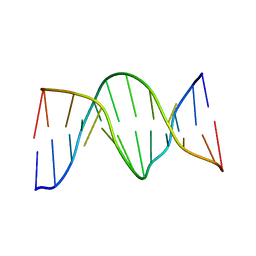 | | Solution Structure of 8OG:G mismatch containing duplex | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(8OG)P*GP*CP*G)-3' | | Authors: | Thiviyanathan, V, Somasunderam, A.D, Hazra, T.K, Mitra, S, Gorenstein, D.G. | | Deposit date: | 2002-10-24 | | Release date: | 2002-11-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a DNA Duplex Containing 8-Hydroxy-2'-Deoxyguanosine Opposite Deoxyguanosine
J.Mol.Biol., 325, 2003
|
|
1K9L
 
 | |
1OZO
 
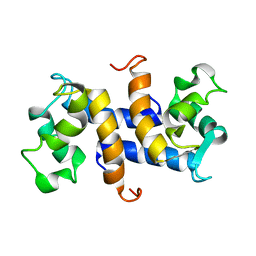 | | Three-dimensional solution structure of apo-S100P protein determined by NMR spectroscopy | | Descriptor: | S-100P protein | | Authors: | Lee, Y.-C, Volk, D.E, Thiviyanathan, V, Kleerekoper, Q, Gribenko, A.V, Zhang, S, Gorenstein, D.G, Makhatadze, G.I, Luxon, B.A. | | Deposit date: | 2003-04-09 | | Release date: | 2004-04-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the Apo-S100P protein.
J.Biomol.Nmr, 29, 2004
|
|
1K9H
 
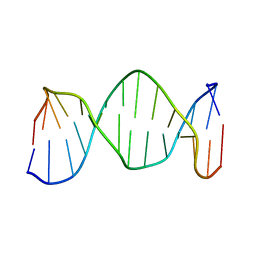 | | NMR structure of DNA TGTGAGCGCTCACA | | Descriptor: | 5'-D(*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*A)-3' | | Authors: | Kaluarachchi, K, Gorenstein, D.G, Luxon, B.A. | | Deposit date: | 2001-10-29 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | How Do Proteins Recognize DNA? Solution Structure and Local Conformational Dynamics of Lac Operators by 2D NMR
J.Biomol.Struct.Dyn., Conversation 11, 2000
|
|
2L9I
 
 | |
1K8S
 
 | | BULGED ADENOSINE IN AN RNA DUPLEX | | Descriptor: | 5'-R(*GP*CP*GP*GP*CP*AP*CP*CP*UP*GP*CP*C)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*UP*GP*CP*CP*GP*C)-3' | | Authors: | Thiviyanathan, V, Guliaev, A.B, Leontis, N.B, Gorenstein, D.G. | | Deposit date: | 2001-10-25 | | Release date: | 2001-11-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of a bulged adenosine base in an RNA duplex by relaxation matrix refinement.
J.Mol.Biol., 300, 2000
|
|
1IDV
 
 | |
1CXR
 
 | |
1R4H
 
 | | NMR Solution structure of the IIIc domain of GB Virus B IRES Element | | Descriptor: | 5'-R(*GP*GP*GP*CP*AP*AP*GP*CP*CP*C)-3' | | Authors: | Kaluarachchi, K, Thiviyanathan, V, Rijinbrand, R, Lemon, S.M, Gorenstein, D.G. | | Deposit date: | 2003-10-06 | | Release date: | 2004-10-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mutational and structural analysis of stem-loop IIIC of the hepatitis C virus and GB virus B internal ribosome entry sites.
J.Mol.Biol., 343, 2004
|
|
1T28
 
 | | High resolution structure of a picornaviral internal cis-acting replication element | | Descriptor: | 34-MER | | Authors: | Thiviyanathan, V, Yang, Y, Kaluarachchi, K, Reynbrand, R, Gorenstein, D.G, Lemon, S.M. | | Deposit date: | 2004-04-20 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution structure of a picornaviral internal cis-acting replication element(cre).
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
