1AN7
 
 | |
3U56
 
 | |
4X9C
 
 | | 1.4A crystal structure of Hfq from Methanococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nikulin, A.D, Tishchenko, S.V, Nikonova, S.V, Murina, V.N, Mihailina, A.O, Lekontseva, N.V. | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization of RNA-binding properties of the archaeal Hfq-like protein from Methanococcus jannaschii.
J. Biomol. Struct. Dyn., 35, 2017
|
|
5H04
 
 | | Crystal structure of an ADP-ribosylating toxin BECa of a novel binary enterotoxin of C. perfringens with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Binary enterotoxin of Clostridium perfringens component a | | Authors: | Kawahara, K, Yonogi, S, Munetomo, R, Oki, H, Yoshida, T, Ohkubo, T, Kumeda, Y, Matsuda, S, Kodama, T, Iida, T, Nakamura, S. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.825 Å) | | Cite: | Crystal structure of the ADP-ribosylating component of BEC, the binary enterotoxin of Clostridium perfringens.
Biochem.Biophys.Res.Commun., 480, 2016
|
|
5NPM
 
 | | CRYSTAL STRUCTURE OF MUTANT RIBOSOMAL PROTEIN TTHL1 LACKING 8 N-TERMINAL RESIDUES IN COMPLEX WITH 80NT 23S RNA FROM THERMUS THERMOPHILUS | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L1, MALONATE ION, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Nevskaya, N.A, Nikonov, S.V, Garber, M.B. | | Deposit date: | 2017-04-17 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | [Influence of Nonconserved Regions of L1 Protuberance of Thermus thermophilus Ribosome on the Affinity of L1 Protein to 23s rRNA].
Mol.Biol.(Moscow), 52, 2018
|
|
5H03
 
 | | Crystal structure of an ADP-ribosylating toxin BECa from C. perfringens | | Descriptor: | Binary enterotoxin of Clostridium perfringens component a | | Authors: | Kawahara, K, Yonogi, S, Munetomo, R, Oki, H, Yoshida, T, Ohkubo, T, Kumeda, Y, Matsuda, S, Kodama, T, Iida, T, Nakamura, S. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of the ADP-ribosylating component of BEC, the binary enterotoxin of Clostridium perfringens.
Biochem.Biophys.Res.Commun., 480, 2016
|
|
3U4M
 
 | | Crystal structure of ribosomal protein tthl1 in complex with 80nt 23s rna from thermus thermophilus | | Descriptor: | 50S ribosomal protein L1, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, NIkonov, S.V. | | Deposit date: | 2011-10-10 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution crystal structure of the isolated ribosomal L1 stalk.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1U1T
 
 | | Hfq protein from Pseudomonas aeruginosa. High-salt crystals | | Descriptor: | Hfq protein | | Authors: | Nikulin, A.D, Stolboushkina, E.A, Perederina, A.A, Vassilieva, I.M, Blaesi, U, Moll, I, Kachalova, G, Yokoyama, S, Vassylyev, D, Garber, M, Nikonov, S.V, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-16 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Pseudomonas aeruginosa Hfq protein.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1U1S
 
 | | Hfq protein from Pseudomonas aeruginosa. Low-salt crystals | | Descriptor: | Hfq protein | | Authors: | Nikulin, A.D, Stolboushkina, E.A, Perederina, I, Vassilieva, I.M, Blaesi, U, Moll, I, Kachalova, G, Vassylyev, D, Yokoyama, S, Garber, M, Nikonov, S.V, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-16 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Pseudomonas aeruginosa Hfq protein.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1SZ6
 
 | | MISTLETOE LECTIN I FROM VISCUM ALBUM. CRYSTAL STRUCTURE AT 2.05 A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AZIDE ION, BETA-GALACTOSIDE SPECIFIC LECTIN I B CHAIN, ... | | Authors: | Gabdoulkhakov, A.G, Guhlistova, N.E, Lyashenko, A.V, Krauspenhaar, R, Stoeva, S, Voelter, W, Nikonov, S.V, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2004-04-04 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Viscum album Mistletoe Lectin I in native state at 2.05 A resolution, comparison of structure active site conformation in ricin and in viscumin
TO BE PUBLISHED
|
|
5AHV
 
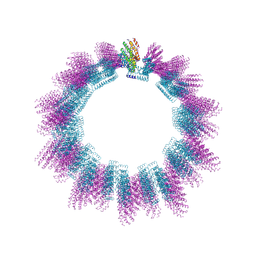 | | Cryo-EM structure of helical ANTH and ENTH tubules on PI(4,5)P2-containing membranes | | Descriptor: | ANTH DOMAIN OF ENDOCYTIC ADAPTOR SLA2, ENTH DOMAIN OF EPSIN ENT1 | | Authors: | Skruzny, M, Desfosses, A, Prinz, S, Dodonova, S.O, Gieras, A, Uetrecht, C, Jakobi, A.J, Abella, M, Hagen, W.J.H, Schulz, J, Meijers, R, Rybin, V, Briggs, J.A.G, Sachse, C, Kaksonen, M. | | Deposit date: | 2015-02-10 | | Release date: | 2015-05-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | An Organized Co-Assembly of Clathrin Adaptors is Essential for Endocytosis.
Dev.Cell, 33, 2015
|
|
