5Z7Q
 
 | | Crystal structure of Bacillus cereus flagellin | | Descriptor: | Flagellin | | Authors: | Kim, M, Hong, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Bacillus cereus flagellin and structure-guided fusion-protein designs
Sci Rep, 8, 2018
|
|
2C8Y
 
 | | thrombin inhibitors | | Descriptor: | DIMETHYL SULFOXIDE, HIRUDIN VARIANT-2, N-[(2R,3S)-3-AMINO-2-HYDROXY-4-PHENYLBUTYL]NAPHTHALENE-2-SULFONAMIDE, ... | | Authors: | Howard, N, Abell, C, Blakemore, W, Carr, R, Chessari, G, Congreve, M, Howard, S, Jhoti, H, Murray, C.W, Seavers, L.C.A, van Montfort, R.L.M. | | Deposit date: | 2005-12-08 | | Release date: | 2006-07-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Application of Fragment Screening and Fragment Linking to the Discovery of Novel Thrombin Inhibitors
J.Med.Chem., 49, 2006
|
|
2XJJ
 
 | | Structure of HSP90 with small molecule inhibitor bound | | Descriptor: | 1,3-DIHYDROISOINDOL-2-YL-(6-HYDROXY-3,3-DIMETHYL-1,2-DIHYDROINDOL-5-YL)METHANONE, GLYCEROL, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, M, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, OBrien, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of (2,4-Dihydroxy-5-Isopropylphenyl)-[5-(4-Methylpiperazin-1-Ylmethyl)-1,3-Dihydroisoindol-2-Yl]Methanone (at13387), a Novel Inhibitor of the Molecular Chaperone Hsp90 by Fragment Based Drug Design.
J.Med.Chem., 53, 2010
|
|
2XHT
 
 | | Structure of HSP90 with small molecule inhibitor bound | | Descriptor: | (3-TERT-BUTYL-4-HYDROXYPHENYL)MORPHOLIN-4-YL-METHANONE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, M, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, OBrien, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-06-21 | | Release date: | 2010-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Fragment-Based Drug Discovery Applied to Hsp90. Discovery of Two Lead Series with High Ligand Efficiency.
J.Med.Chem., 53, 2010
|
|
2XK2
 
 | | Structure of HSP90 with small molecule inhibitor bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK PROTEIN HSP 90-ALPHA, MAGNESIUM ION | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, M, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, OBrien, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-07-07 | | Release date: | 2010-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-Based Drug Discovery Applied to Hsp90. Discovery of Two Lead Series with High Ligand Efficiency.
J.Med.Chem., 53, 2010
|
|
2XDX
 
 | | Structure of HSP90 with small molecule inhibitor bound | | Descriptor: | 4-CHLORO-6-(2-METHOXYPHENYL)PYRIMIDIN-2-AMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, M, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, O'Brien, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Fragment-Based Drug Discovery Applied to Hsp90. Discovery of Two Lead Series with High Ligand Efficiency.
J.Med.Chem., 53, 2010
|
|
2XJX
 
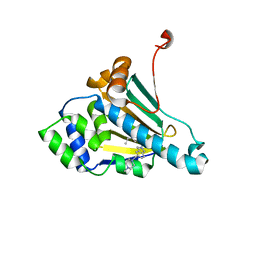 | | Structure of HSP90 with small molecule inhibitor bound | | Descriptor: | HEAT SHOCK PROTEIN HSP 90-ALPHA, Onalespib | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, M, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, OBrien, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of (2,4-Dihydroxy-5-Isopropylphenyl)-[5-(4-Methylpiperazin-1-Ylmethyl)-1,3-Dihydroisoindol-2-Yl]Methanone (at13387), a Novel Inhibitor of the Molecular Chaperone Hsp90 by Fragment Based Drug Design.
J.Med.Chem., 53, 2010
|
|
2XAB
 
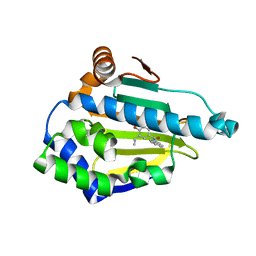 | | Structure of HSP90 with an inhibitor bound | | Descriptor: | 4-(1,3-DIHYDRO-2H-ISOINDOL-2-YLCARBONYL)-6-(1-METHYLETHYL)BENZENE-1,3-DIOL, HEAT SHOCK PROTEIN HSP 90-ALPHA, | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, m, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, O'Brian, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-03-30 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of (2,4-Dihydroxy-5-Isopropylphenyl)-[5-(4-Methylpiperazin-1-Ylmethyl)-1,3-Dihydroisoindol-2-Yl]Methanone (at13387), a Novel Inhibitor of the Molecular Chaperone Hsp90 by Fragment Based Drug Design.
J.Med.Chem., 53, 2010
|
|
2XDK
 
 | | Structure of HSP90 with small molecule inhibitor bound | | Descriptor: | 2-AMINO-4-PYRIDYL-PYRIMIDINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, M, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, OBrien, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-05-04 | | Release date: | 2010-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Fragment-Based Drug Discovery Applied to Hsp90. Discovery of Two Lead Series with High Ligand Efficiency.
J.Med.Chem., 53, 2010
|
|
5ZQH
 
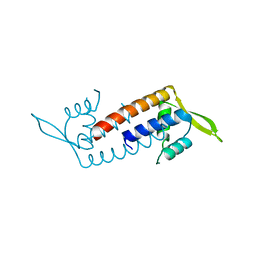 | | Crystal structure of Streptococcus transcriptional regulator | | Descriptor: | PadR family transcriptional regulator | | Authors: | Kim, M, Hong, M. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based functional analysis of a PadR transcription factor from Streptococcus pneumoniae and characteristic features in the PadR subfamily-2.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
5A8E
 
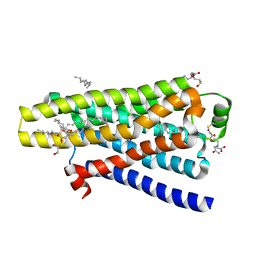 | | thermostabilised beta1-adrenoceptor with rationally designed inverse agonist 7-methylcyanopindolol bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 4-[(2S)-3-(tert-butylamino)-2-hydroxypropoxy]-7-methyl-1H-indole-2-carbonitrile, ... | | Authors: | Sato, T, Baker, J.G, Warne, T, Brown, G.A, Congreve, M, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2015-07-15 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pharmacological Analysis and Structure Determination of 7-Methylcyanopindolol-Bound Beta1-Adrenergic Receptor.
Mol.Pharmacol., 88, 2015
|
|
5B6A
 
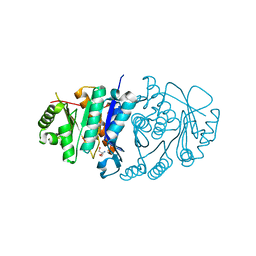 | | Structure of Pyridoxal Kinasefrom Pseudomonas Aeruginosa | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kim, M.I, Hong, M. | | Deposit date: | 2016-05-25 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and catalytic mechanism of pyridoxal kinase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 478, 2016
|
|
8G55
 
 | |
8G54
 
 | |
4ISR
 
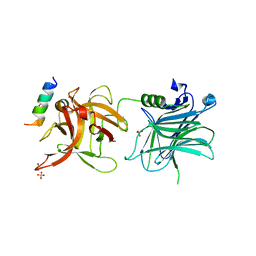 | | Binding domain of Botulinum neurotoxin DC in complex with rat synaptotagmin II | | Descriptor: | Neurotoxin, SULFATE ION, Synaptotagmin-2 | | Authors: | Berntsson, R.P.-A, Peng, L, Svensson, L.M, Dong, M, Stenmark, P. | | Deposit date: | 2013-01-17 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structures of Botulinum Neurotoxin DC in Complex with Its Protein Receptors Synaptotagmin I and II.
Structure, 21, 2013
|
|
1OEV
 
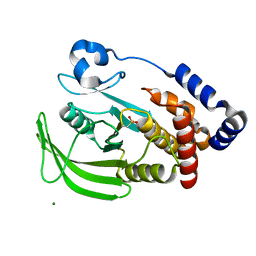 | | Oxidation state of protein tyrosine phosphatase 1B | | Descriptor: | MAGNESIUM ION, PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | van Montfort, R.L.M, Congreve, M, Tisi, D, Carr, R, Jhoti, H. | | Deposit date: | 2003-03-31 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B.
Nature, 423, 2003
|
|
4ISQ
 
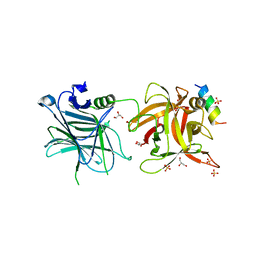 | | Binding domain of Botulinum neurotoxin DC in complex with human synaptotagmin I | | Descriptor: | GLYCEROL, Neurotoxin, SULFATE ION, ... | | Authors: | Berntsson, R.P.-A, Peng, L, Svensson, L.M, Dong, M, Stenmark, P. | | Deposit date: | 2013-01-17 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of Botulinum Neurotoxin DC in Complex with Its Protein Receptors Synaptotagmin I and II.
Structure, 21, 2013
|
|
7DVD
 
 | | The crystal structure of p53 DNA binding domain and PUMA complex | | Descriptor: | Bcl-2-binding component 3, isoforms 1/2, Cellular tumor antigen p53, ... | | Authors: | Han, C.W, Lee, H.N, Jeong, M.S, Jang, S.B. | | Deposit date: | 2021-01-13 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis of the p53 DNA binding domain and PUMA complex.
Biochem.Biophys.Res.Commun., 548, 2021
|
|
1OET
 
 | | Oxidation state of protein tyrosine phosphatase 1B | | Descriptor: | -TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1, MAGNESIUM ION | | Authors: | van Montfort, R.L.M, Congreve, M, Tisi, D, Carr, R, Jhoti, H. | | Deposit date: | 2003-03-31 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B.
Nature, 423, 2003
|
|
2M35
 
 | | NMR study of k-Ssm1a | | Descriptor: | k-Ssm1a | | Authors: | King, G.F, Undheim, E.A, Mobli, M, Yang, S, Rong, M, Lai, R. | | Deposit date: | 2013-01-09 | | Release date: | 2014-01-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR study of k-Ssm1a
To be Published
|
|
1OES
 
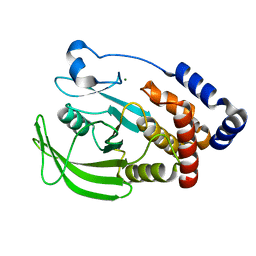 | | Oxidation state of protein tyrosine phosphatase 1B | | Descriptor: | MAGNESIUM ION, PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | van Montfort, R.L.M, Congreve, M, Tisi, D, Carr, R, Jhoti, H. | | Deposit date: | 2003-03-31 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B.
Nature, 423, 2003
|
|
1OEU
 
 | | Oxidation state of protein tyrosine phosphatase 1B | | Descriptor: | MAGNESIUM ION, PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | van Montfort, R.L.M, Congreve, M, Tisi, D, Carr, R, Jhoti, H. | | Deposit date: | 2003-03-31 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B.
Nature, 423, 2003
|
|
6ABT
 
 | | Crystal structure of transcription factor from Listeria monocytogenes | | Descriptor: | PadR family transcriptional regulator | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based molecular characterization and regulatory mechanism of the LftR transcription factor from Listeria monocytogenes: Conformational flexibilities and a ligand-induced regulatory mechanism.
Plos One, 14, 2019
|
|
5XEF
 
 | | Crystal structure of flagellar chaperone from bacteria | | Descriptor: | Flagellar protein fliS | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2017-04-05 | | Release date: | 2018-06-27 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the flagellar chaperone FliS from Bacillus cereus and an invariant proline critical for FliS dimerization and flagellin recognition
Biochem. Biophys. Res. Commun., 487, 2017
|
|
6ABQ
 
 | | Crystal structure of transcription factor from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, PadR family transcriptional regulator | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based molecular characterization and regulatory mechanism of the LftR transcription factor from Listeria monocytogenes: Conformational flexibilities and a ligand-induced regulatory mechanism.
Plos One, 14, 2019
|
|
