1BR4
 
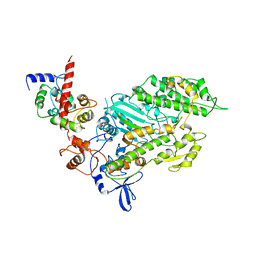 | | SMOOTH MUSCLE MYOSIN MOTOR DOMAIN-ESSENTIAL LIGHT CHAIN COMPLEX WITH MGADP.BEF3 BOUND AT THE ACTIVE SITE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Dominguez, R, Trybus, K.M, Cohen, C. | | Deposit date: | 1998-08-27 | | Release date: | 1998-09-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state.
Cell(Cambridge,Mass.), 94, 1998
|
|
1BR1
 
 | | SMOOTH MUSCLE MYOSIN MOTOR DOMAIN-ESSENTIAL LIGHT CHAIN COMPLEX WITH MGADP.ALF4 BOUND AT THE ACTIVE SITE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN, ... | | Authors: | Dominguez, R, Trybus, K.M, Cohen, C. | | Deposit date: | 1998-08-26 | | Release date: | 1998-09-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state.
Cell(Cambridge,Mass.), 94, 1998
|
|
2AO5
 
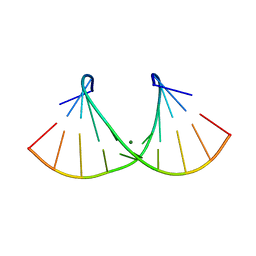 | | Crystal structure of an RNA duplex r(GGCGBrUGCGCU)2 with terminal and internal tandem G-U base pairs | | Descriptor: | 5'-R(*GP*GP*CP*GP*(5BU)P*GP*CP*GP*CP*U)-3', MAGNESIUM ION | | Authors: | Utsunomiya, R, Suto, K, Balasundaresan, D, Fukamizu, A, Kumar, P.K, Mizuno, H. | | Deposit date: | 2005-08-12 | | Release date: | 2006-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of an RNA duplex r(GGCGBrUGCGCU)2 with terminal and internal tandem G.U base pairs.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2MZ0
 
 | |
3WJ8
 
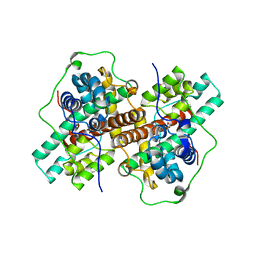 | | Crystal Structure of DL-2-haloacid dehalogenase mutant with 2-bromo-2-methylpropionate | | Descriptor: | 2-bromo-2-methylpropanoic acid, DL-2-haloacid dehalogenase, GLYCEROL | | Authors: | Siwek, A, Omi, R, Hirotsu, K, Jitsumori, K, Esaki, N, Kurihara, T, Paneth, P. | | Deposit date: | 2013-10-07 | | Release date: | 2013-11-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding modes of DL-2-haloacid dehalogenase revealed by crystallography, modeling and isotope effects studies.
Arch.Biochem.Biophys., 540, 2013
|
|
2YRR
 
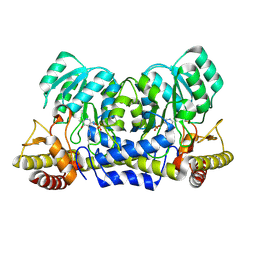 | | hypothetical alanine aminotransferase (TTH0173) from Thermus thermophilus HB8 | | Descriptor: | Aminotransferase, class V, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Miyahara, I, Matsumura, M, Goto, M, Omi, R, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | hypothetical alanine aminotransferase (TTH0173) from Thermus thermophilus HB8
To be Published
|
|
2YRI
 
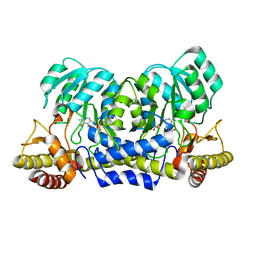 | | Crystal structure of alanine-pyruvate aminotransferase with 2-methylserine | | Descriptor: | (S,E)-3-HYDROXY-2-((3-HYDROXY-2-METHYL-5-(PHOSPHONOOXYMETHYL)PYRIDIN-4-YL)METHYLENEAMINO)-2-METHYLPROPANOIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase, ... | | Authors: | Miyahara, I, Matsumura, M, Goto, M, Omi, R, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | hypothetical alanine aminotransferase (TTHA0173) from Thermus thermophilus HB8
To be Published
|
|
4N2X
 
 | | Crystal Structure of DL-2-haloacid dehalogenase | | Descriptor: | DL-2-haloacid dehalogenase, GLYCEROL | | Authors: | Siwek, A, Omi, R, Hirotsu, K, Jitsumori, K, Esaki, N, Kurihara, T, Paneth, P. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding modes of DL-2-haloacid dehalogenase revealed by crystallography, modeling and isotope effects studies.
Arch.Biochem.Biophys., 540, 2013
|
|
4Z35
 
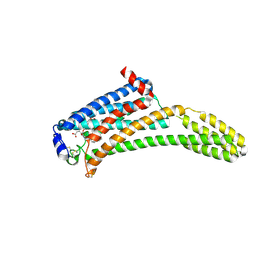 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO-9910539 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 3-{1-[(2S,3S)-3-(4-acetyl-3,5-dimethoxyphenyl)-2-(2,3-dihydro-1H-inden-2-ylmethyl)-3-hydroxypropyl]-4-(methoxycarbonyl)-1H-pyrrol-3-yl}propanoic acid, Lysophosphatidic acid receptor 1,Soluble cytochrome b562 | | Authors: | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | Deposit date: | 2015-03-30 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
4Z36
 
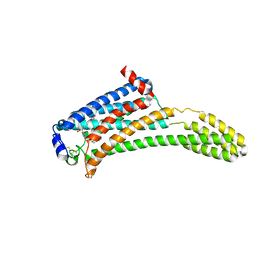 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO-3080573 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 1-(4-{[(2S,3R)-2-(2,3-dihydro-1H-inden-2-yloxy)-3-(3,5-dimethoxy-4-methylphenyl)-3-hydroxypropyl]oxy}phenyl)cyclopropanecarboxylic acid, Lysophosphatidic acid receptor 1,Soluble cytochrome b562 | | Authors: | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | Deposit date: | 2015-03-30 | | Release date: | 2015-06-03 | | Last modified: | 2015-07-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
4Z34
 
 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO9780307 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, Lysophosphatidic acid receptor 1, Soluble cytochrome b562, ... | | Authors: | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | Deposit date: | 2015-03-30 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
1WKG
 
 | | Acetylornithine aminotransferase from thermus thermophilus HB8 | | Descriptor: | Acetylornithine/acetyl-lysine aminotransferase, N~2~-ACETYL-N~5~-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-ORNITHINE | | Authors: | Matsumura, M, Goto, M, Omi, R, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-31 | | Release date: | 2005-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Acetylornithine aminotransferase from thermus thermophilus HB8
To be Published
|
|
1WKH
 
 | | Acetylornithine aminotransferase from thermus thermophilus HB8 | | Descriptor: | 4-[(1,3-DICARBOXY-PROPYLAMINO)-METHYL]-3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDINIUM, Acetylornithine/acetyl-lysine aminotransferase | | Authors: | Matsumura, M, Goto, M, Omi, R, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-31 | | Release date: | 2005-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Acetylornithine aminotransferase from thermus thermophilus HB8
To be Published
|
|
1VEF
 
 | | Acetylornithine aminotransferase from Thermus thermophilus HB8 | | Descriptor: | Acetylornithine/acetyl-lysine aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matsumura, M, Goto, M, Omi, R, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-30 | | Release date: | 2005-08-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Three-Dimensional Strutcure of Acetylornithine aminotransferase from Thermus thermophilus HB8
To be Published
|
|
2CWH
 
 | | Crystal Structure of delta1-piperideine-2-carboxylate reductase from Pseudomonas syringae complexed with NADPH and pyrrole-2-carboxylate | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRROLE-2-CARBOXYLATE, delta1-piperideine-2-carboxylate reductase | | Authors: | Goto, M, Muramatsu, H, Mihara, H, Kurihara, T, Esaki, N, Omi, R, Miyahara, I, Hirotsu, K. | | Deposit date: | 2005-06-20 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of Delta1-piperideine-2-carboxylate/Delta1-pyrroline-2-carboxylate reductase belonging to a new family of NAD(P)H-dependent oxidoreductases: conformational change, substrate recognition, and stereochemistry of the reaction
J.Biol.Chem., 280, 2005
|
|
2CWF
 
 | | Crystal Structure of delta1-piperideine-2-carboxylate reductase from Pseudomonas syringae complexed with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, delta1-piperideine-2-carboxylate reductase | | Authors: | Goto, M, Muramatsu, H, Mihara, H, Kurihara, T, Esaki, N, Omi, R, Miyahara, I, Hirotsu, K. | | Deposit date: | 2005-06-20 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of Delta1-piperideine-2-carboxylate/Delta1-pyrroline-2-carboxylate reductase belonging to a new family of NAD(P)H-dependent oxidoreductases: conformational change, substrate recognition, and stereochemistry of the reaction
J.Biol.Chem., 280, 2005
|
|
1WTJ
 
 | | Crystal Structure of delta1-piperideine-2-carboxylate reductase from Pseudomonas syringae pvar.tomato | | Descriptor: | ureidoglycolate dehydrogenase | | Authors: | Goto, M, Muramatsu, H, Mihara, H, Kurihara, T, Esaki, N, Omi, R, Miyahara, I, Hirotsu, K. | | Deposit date: | 2004-11-24 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of Delta1-piperideine-2-carboxylate/Delta1-pyrroline-2-carboxylate reductase belonging to a new family of NAD(P)H-dependent oxidoreductases: conformational change, substrate recognition, and stereochemistry of the reaction
J.Biol.Chem., 280, 2005
|
|
1K8U
 
 | |
1K96
 
 | | CRYSTAL STRUCTURE OF CALCIUM BOUND HUMAN S100A6 | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, S100A6 | | Authors: | Otterbein, L.R, Dominguez, R. | | Deposit date: | 2001-10-26 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structures of S100A6 in the Ca(2+)-free and Ca(2+)-bound states: the calcium sensor mechanism of S100 proteins revealed at atomic resolution.
Structure, 10, 2002
|
|
1K9K
 
 | | CRYSTAL STRUCTURE OF CALCIUM BOUND HUMAN S100A6 | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, S100A6 | | Authors: | Otterbein, L.R, Dominguez, R. | | Deposit date: | 2001-10-29 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structures of S100A6 in the Ca(2+)-free and Ca(2+)-bound states: the calcium sensor mechanism of S100 proteins revealed at atomic resolution.
Structure, 10, 2002
|
|
1K9P
 
 | |
1UG9
 
 | | Crystal Structure of Glucodextranase from Arthrobacter globiformis I42 | | Descriptor: | CALCIUM ION, GLYCEROL, glucodextranase | | Authors: | Mizuno, M, Tonozuka, T, Suzuki, S, Uotsu-Tomita, R, Ohtaki, A, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2003-06-16 | | Release date: | 2003-12-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into substrate specificity and function of glucodextranase
J.Biol.Chem., 279, 2004
|
|
1CEC
 
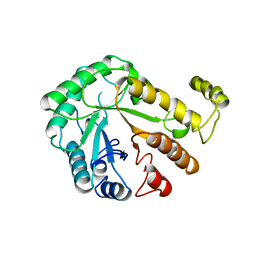 | |
1TN4
 
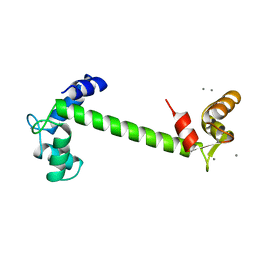 | | FOUR CALCIUM TNC | | Descriptor: | CALCIUM ION, TROPONIN C | | Authors: | Love, M.L, Dominguez, R, Houdusse, A, Cohen, C. | | Deposit date: | 1997-09-18 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of four Ca2+-bound troponin C at 2.0 A resolution: further insights into the Ca2+-switch in the calmodulin superfamily.
Structure, 5, 1997
|
|
9B0K
 
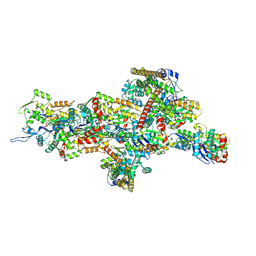 | | INF2 in the Middle of F-Actin (Down state) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Palmer, N.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2024-03-12 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Mechanisms of actin filament severing and elongation by formins.
Nature, 632, 2024
|
|
