7H5K
 
 | | Crystal structure of endothiapepsin IS_RT2 in complex with AC40075 at 296 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.027 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5O
 
 | | Crystal structure of endothiapepsin PF_cryo3 in complex with TL00150 at 100 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5R
 
 | | Crystal structure of endothiapepsin IS_cryo3 in complex with TL00150 at 100 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H56
 
 | | Crystal structure of endothiapepsin PF_cryo3 in complex with AC39729 at 100 K | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.987 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5I
 
 | | Crystal structure of endothiapepsin IS_cryo3 in complex with AC40075 at 100 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5A
 
 | | Crystal structure of endothiapepsin IS_cryo1 in complex with AC39729 at 100 K | | Descriptor: | 5-fluoranylpyridin-2-amine, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5D
 
 | | Crystal structure of endothiapepsin PN_cryo1 in complex with AC40075 at 100 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.907 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5J
 
 | | Crystal structure of endothiapepsin IS_cryo1 in complex with AC40075 at 100 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5N
 
 | | Crystal structure of endothiapepsin PN_RT2 in complex with TL00150 at 296 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.857 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5Q
 
 | | Crystal structure of endothiapepsin PF_RT2 in complex with TL00150 at 296 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5T
 
 | | Crystal structure of endothiapepsin IS_RT2 in complex with TL00150 at 296 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6ZRW
 
 | | Crystal structure of the fungal lectin CML1 | | Descriptor: | ACETATE ION, GLYCEROL, Mucin-binding lectin 1, ... | | Authors: | Bleuler-Martinez, S, Olieric, V, Sharpe, M, Capitani, G, Aebi, M, Kuenzler, M. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-function relationship of a novel fucoside-binding fruiting body lectin from Coprinopsis cinerea exhibiting nematotoxic activity.
Glycobiology, 32, 2022
|
|
7B0O
 
 | | In meso structure of the membrane integral lipoprotein intramolecular transacylase Lit from Bacillus cereus in space group P21 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Huang, C.-Y, Olatunji, S, Olieric, V, Caffrey, M. | | Deposit date: | 2020-11-20 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural basis of the membrane intramolecular transacylase reaction responsible for lyso-form lipoprotein synthesis.
Nat Commun, 12, 2021
|
|
7B0P
 
 | | In meso structure of the membrane integral lipoprotein intramolecular transacylase Lit from Bacillus cereus in space group P21212 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, GLYCEROL, ... | | Authors: | Huang, C.-Y, Olatunji, S, Olieric, V, Caffrey, M. | | Deposit date: | 2020-11-20 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Structural basis of the membrane intramolecular transacylase reaction responsible for lyso-form lipoprotein synthesis.
Nat Commun, 12, 2021
|
|
7B0R
 
 | | In meso structure of the membrane integral lipoprotein intramolecular transacylase Lit from Bacillus cereus with H85R mutation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Huang, C.-Y, Olatunji, S, Olieric, V, Caffrey, M. | | Deposit date: | 2020-11-20 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the membrane intramolecular transacylase reaction responsible for lyso-form lipoprotein synthesis.
Nat Commun, 12, 2021
|
|
7B0Q
 
 | | In meso structure of the membrane integral lipoprotein intramolecular transacylase Lit from Bacillus cereus with H85A mutation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, CITRIC ACID, ... | | Authors: | Huang, C.-Y, Olatunji, S, Olieric, V, Caffrey, M. | | Deposit date: | 2020-11-20 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis of the membrane intramolecular transacylase reaction responsible for lyso-form lipoprotein synthesis.
Nat Commun, 12, 2021
|
|
7AZ5
 
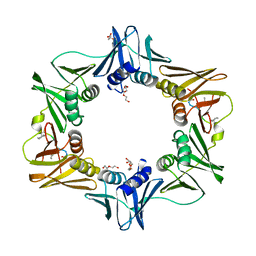 | | DNA polymerase sliding clamp from Escherichia coli with peptide 47 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, Peptide 47, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZG
 
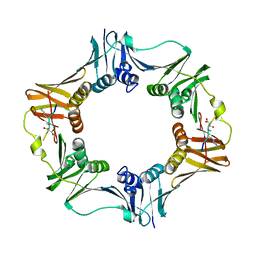 | | DNA polymerase sliding clamp from Escherichia coli with peptide 4 bound | | Descriptor: | Beta sliding clamp, Peptide 4 | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZL
 
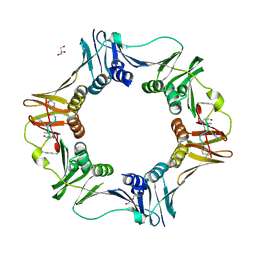 | | DNA polymerase sliding clamp from Escherichia coli with peptide 38 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZ7
 
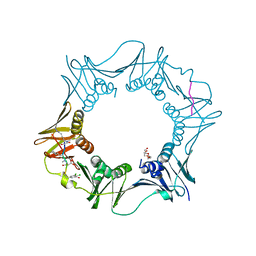 | | DNA polymerase sliding clamp from Escherichia coli with peptide 37 bound | | Descriptor: | Beta sliding clamp, FORMIC ACID, PENTAETHYLENE GLYCOL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZ6
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 36 bound | | Descriptor: | ACETATE ION, Beta sliding clamp, CHLORIDE ION, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZ8
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 43 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZD
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 20 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZE
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 18 bound | | Descriptor: | Beta sliding clamp, GLYCEROL, MALONATE ION, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZC
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 22 bound | | Descriptor: | Beta sliding clamp, GLYCEROL, Peptide 22 | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
