1D8J
 
 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
8I53
 
 | |
2DY7
 
 | |
2DY8
 
 | |
5GOW
 
 | |
1D8K
 
 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
5XV8
 
 | |
7BUL
 
 | |
2RNQ
 
 | |
2RNR
 
 | |
2RVB
 
 | |
2RUK
 
 | |
6K5W
 
 | | Solution structure of the chromodomain of yeast Eaf3 | | Descriptor: | Chromatin modification-related protein EAF3 | | Authors: | Okuda, M, Nishimura, Y. | | Deposit date: | 2019-05-31 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Eaf3 chromodomain acts as a pH sensor for gene expression by altering its binding affinity for histone methylated-lysine residues.
Biosci.Rep., 40, 2020
|
|
7DTI
 
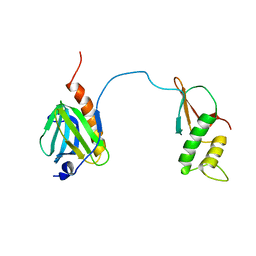 | |
7DTH
 
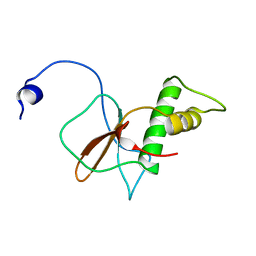 | | Solution structure of RPB6, common subunit of RNA polymerases I, II, and III | | Descriptor: | DNA-directed RNA polymerases I, II, and III subunit RPABC2 | | Authors: | Okuda, M, Nishimura, Y. | | Deposit date: | 2021-01-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Three human RNA polymerases interact with TFIIH via a common RPB6 subunit.
Nucleic Acids Res., 50, 2022
|
|
1VD4
 
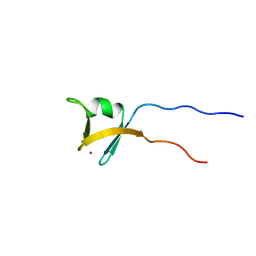 | | Solution structure of the zinc finger domain of TFIIE alpha | | Descriptor: | Transcription initiation factor IIE, alpha subunit, ZINC ION | | Authors: | Okuda, M, Tanaka, A, Arai, Y, Satoh, M, Okamura, H, Nagadoi, A, Hanaoka, F, Ohkuma, Y, Nishimura, Y. | | Deposit date: | 2004-03-18 | | Release date: | 2004-10-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel zinc finger structure in the large subunit of human general transcription factor TFIIE.
J.Biol.Chem., 279, 2004
|
|
6SEV
 
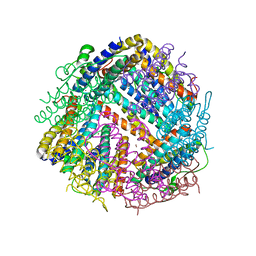 | |
3AJV
 
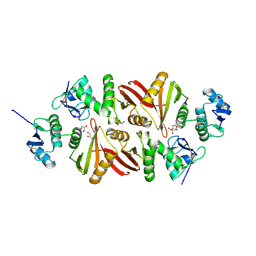 | | Splicing endonuclease from Aeropyrum pernix | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative uncharacterized protein, ... | | Authors: | Yoshinari, S, Watanabe, Y, Okuda, M, Shiba, T, Inaoka, K.D, Kurisu, G. | | Deposit date: | 2010-06-19 | | Release date: | 2010-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Conserved Lysine Residue in the Crenarchaea-Specific Loop is Important for the Crenarchaeal Splicing Endonuclease Activity.
J.Mol.Biol., 405, 2011
|
|
2RO0
 
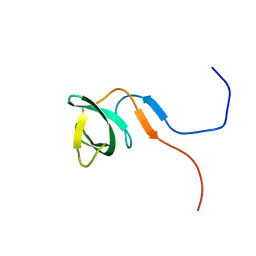 | | Solution structure of the knotted tudor domain of the yeast histone acetyltransferase, Esa1 | | Descriptor: | Histone acetyltransferase ESA1 | | Authors: | Shimojo, H, Sano, N, Moriwaki, Y, Okuda, M, Horikoshi, M, Nishimura, Y. | | Deposit date: | 2008-03-01 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Novel structural and functional mode of a knot essential for RNA binding activity of the Esa1 presumed chromodomain
J.Mol.Biol., 378, 2008
|
|
2RNZ
 
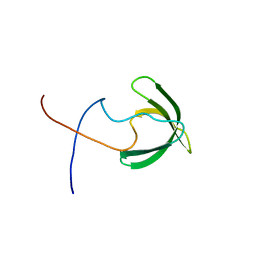 | | Solution structure of the presumed chromodomain of the yeast histone acetyltransferase, Esa1 | | Descriptor: | Histone acetyltransferase ESA1 | | Authors: | Shimojo, H, Sano, N, Moriwaki, Y, Okuda, M, Horikoshi, M, Nishimura, Y. | | Deposit date: | 2008-03-01 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Novel structural and functional mode of a knot essential for RNA binding activity of the Esa1 presumed chromodomain
J.Mol.Biol., 378, 2008
|
|
6HX2
 
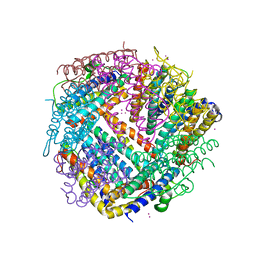 | |
6HUI
 
 | |
6HVQ
 
 | |
6HV1
 
 | |
