2DBX
 
 | | Crystal Structure of Gamma-glutamyltranspeptidase from Escherichia coli Complexed with L-Glutamate | | Descriptor: | CALCIUM ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Okada, T, Wada, K, Fukuyama, K. | | Deposit date: | 2005-12-16 | | Release date: | 2006-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of gamma-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DBU
 
 | |
2DG5
 
 | | Crystal Structure of Gamma-glutamyl transpeptidase from Escherichia coli in complex with hydrolyzed Glutathione | | Descriptor: | CALCIUM ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Okada, T, Wada, K, Fukuyama, K. | | Deposit date: | 2006-03-08 | | Release date: | 2006-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of gamma-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2ZXJ
 
 | |
7XJV
 
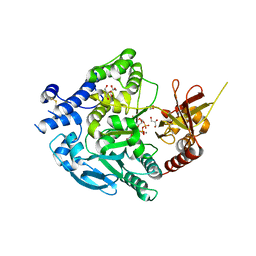 | | Crystal Structure of Alpha-1,3-mannosyltransferase MNT2 from Saccharomyces cerevisiae, Mn/GDP-mannose form | | Descriptor: | Alpha-1,3-mannosyltransferase MNT2, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | Authors: | Hira, D, Kadooka, C, Oka, T. | | Deposit date: | 2022-04-18 | | Release date: | 2023-05-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Alpha-1,3-mannosyltransferase MNT2 from Saccharomyces cerevisiae, Mn/GDP-mannose form
To Be Published
|
|
7BOO
 
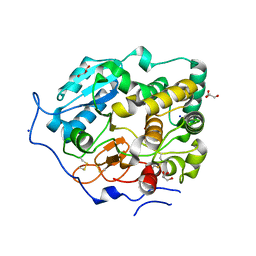 | | Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, apo form | | Descriptor: | Alpha-1,2-mannosyltransferase (Ktr4), putative, GLYCEROL, ... | | Authors: | Hira, D, Onoue, T, Oka, T. | | Deposit date: | 2020-03-19 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the core-mannan biosynthesis of cell wall fungal-type galactomannan in Aspergillus fumigatus .
J.Biol.Chem., 295, 2020
|
|
7BOP
 
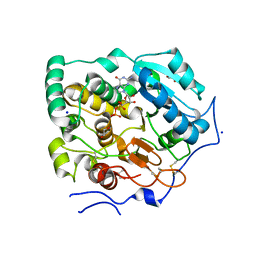 | | Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, Mn/GDP-form | | Descriptor: | Alpha-1,2-mannosyltransferase (Ktr4), putative, GLYCEROL, ... | | Authors: | Hira, D, Onoue, T, Oka, T. | | Deposit date: | 2020-03-19 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the core-mannan biosynthesis of cell wall fungal-type galactomannan in Aspergillus fumigatus .
J.Biol.Chem., 295, 2020
|
|
1QAH
 
 | |
7DOD
 
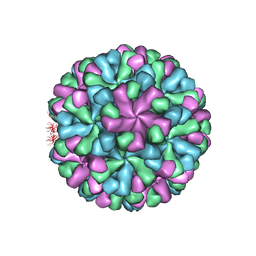 | | Capsid structure of human sapovirus | | Descriptor: | Calicivirin | | Authors: | Miyazaki, N, Murakami, K, Oka, T, Iwasaki, K, Katayama, K, Murata, K. | | Deposit date: | 2020-12-14 | | Release date: | 2021-12-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Atomic structure of human sapovirus capsid by single particle cryo-electron microscopy
To Be Published
|
|
1WZ1
 
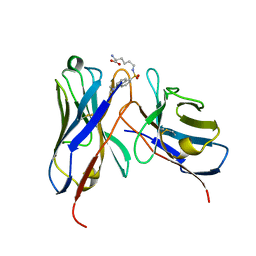 | | Crystal structure of the Fv fragment complexed with dansyl-lysine | | Descriptor: | Ig heavy chain, Ig light chain, N~6~-{[5-(DIMETHYLAMINO)-1-NAPHTHYL]SULFONYL}-L-LYSINE | | Authors: | Nakasako, M, Oka, T, Mashumo, M, Takahashi, H, Shimada, I, Yamaguchi, Y, Kato, K, Arata, Y. | | Deposit date: | 2005-02-21 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational dynamics of complementarity-determining region H3 of an anti-dansyl Fv fragment in the presence of its hapten
J.Mol.Biol., 351, 2005
|
|
1X1S
 
 | | Crystal structure of M-Ras in complex with GppNHp | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein M-Ras | | Authors: | Ye, M, Shima, F, Muraoka, S, Liao, J, Okamoto, H, Yamamoto, M, Tamura, A, Yagi, N, Ueki, T, Kataoka, T. | | Deposit date: | 2005-04-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of M-Ras Reveals a GTP-bound "Off" State Conformation of Ras Family Small GTPases
J.Biol.Chem., 280, 2005
|
|
1X1R
 
 | | Crystal structure of M-Ras in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein M-Ras | | Authors: | Ye, M, Shima, F, Muraoka, S, Liao, J, Okamoto, H, Yamamoto, M, Tamura, A, Yagi, N, Ueki, T, Kataoka, T. | | Deposit date: | 2005-04-12 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of M-Ras Reveals a GTP-bound "Off" State Conformation of Ras Family Small GTPases
J.Biol.Chem., 280, 2005
|
|
8JJE
 
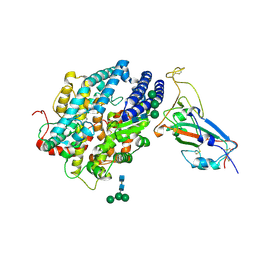 | | RBD of SARS-CoV2 spike protein with ACE2 decoy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Kishikawa, J, Hirose, M, Kato, T, Okamoto, T. | | Deposit date: | 2023-05-30 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An inhaled ACE2 decoy confers protection against SARS-CoV-2 infection in preclinical models.
Sci Transl Med, 15, 2023
|
|
7WXS
 
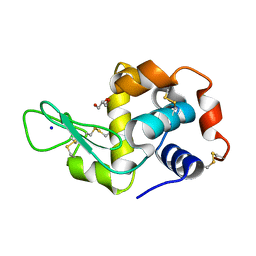 | | Lysozyme protected by polyacrylamide gel | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Muroyama, H, Tomoike, F, Nagae, T, Okada, T. | | Deposit date: | 2022-02-15 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Post-crystallization protection of protein crystals by polyacrylamide
To Be Published
|
|
7WXT
 
 | |
8J6G
 
 | |
1IQZ
 
 | | OXIDIZED [4Fe-4S] FERREDOXIN FROM BACILLUS THERMOPROTEOLYTICUS (FORM I) | | Descriptor: | Ferredoxin, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Fukuyama, K, Okada, T, Kakuta, Y, Takahashi, Y. | | Deposit date: | 2001-08-30 | | Release date: | 2002-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Atomic resolution structures of oxidized [4Fe-4S] ferredoxin from Bacillus thermoproteolyticus in two crystal forms: systematic distortion of [4Fe-4S] cluster in the protein.
J.Mol.Biol., 315, 2002
|
|
1IR0
 
 | | OXIDIZED [4Fe-4S] FERREDOXIN FROM BACILLUS THERMOPROTEOLYTICUS (FORM II) | | Descriptor: | Ferredoxin, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Fukuyama, K, Okada, T, Kakuta, Y, Takahashi, Y. | | Deposit date: | 2001-08-30 | | Release date: | 2002-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution structures of oxidized [4Fe-4S] ferredoxin from Bacillus thermoproteolyticus in two crystal forms: systematic distortion of [4Fe-4S] cluster in the protein.
J.Mol.Biol., 315, 2002
|
|
1IUF
 
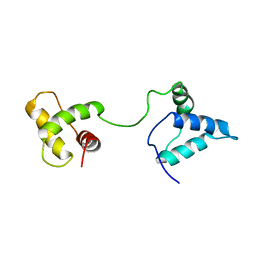 | | LOW RESOLUTION SOLUTION STRUCTURE OF THE TWO DNA-BINDING DOMAINS IN Schizosaccharomyces pombe ABP1 PROTEIN | | Descriptor: | centromere abp1 protein | | Authors: | Kikuchi, J, Iwahara, J, Kigawa, T, Murakami, Y, Okazaki, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-04 | | Release date: | 2002-06-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure determination of the two DNA-binding domains in the Schizosaccharomyces pombe Abp1 protein by a combination of dipolar coupling and diffusion anisotropy restraints.
J.Biomol.NMR, 22, 2002
|
|
2G87
 
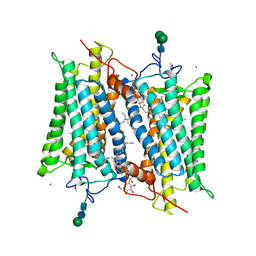 | | Crystallographic model of bathorhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Nakamichi, H, Okada, T. | | Deposit date: | 2006-03-02 | | Release date: | 2006-09-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic analysis of primary visual photochemistry
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
3KKQ
 
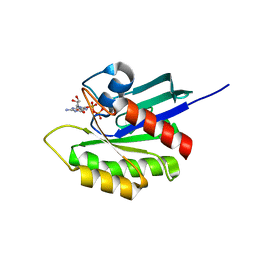 | | Crystal structure of M-Ras P40D in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein M-Ras | | Authors: | Muraoka, S, Shima, F, Liao, J, Ijiri, Y, Matsumoto, K, Ye, M, Inoue, T, Kataoka, T. | | Deposit date: | 2009-11-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for conformational dynamics of GTP-bound Ras protein
J.Biol.Chem., 285, 2010
|
|
3KKM
 
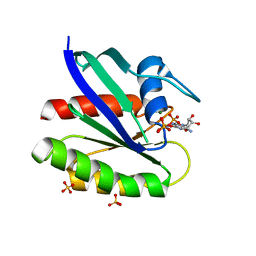 | | Crystal structure of H-Ras T35S in complex with GppNHp | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Muraoka, S, Shima, F, Liao, J, Ijiri, Y, Matsumoto, K, Ye, M, Inoue, T, Kataoka, T. | | Deposit date: | 2009-11-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for conformational dynamics of GTP-bound Ras protein
J.Biol.Chem., 285, 2010
|
|
3KKP
 
 | | Crystal structure of M-Ras P40D in complex with GppNHp | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein M-Ras | | Authors: | Muraoka, S, Shima, F, Liao, J, Ijiri, Y, Matsumoto, K, Ye, M, Inoue, T, Kataoka, T. | | Deposit date: | 2009-11-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for conformational dynamics of GTP-bound Ras protein
J.Biol.Chem., 285, 2010
|
|
3KKO
 
 | | Crystal structure of M-Ras P40D/D41E/L51R in complex with GppNHp | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein M-Ras, ... | | Authors: | Muraoka, S, Shima, F, Liao, J, Ijiri, Y, Matsumoto, K, Ye, M, Inoue, T, Kataoka, T. | | Deposit date: | 2009-11-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for conformational dynamics of GTP-bound Ras protein
J.Biol.Chem., 285, 2010
|
|
3KKN
 
 | | Crystal structure of H-Ras T35S in complex with GppNHp | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Muraoka, S, Shima, F, Liao, J, Ijiri, Y, Matsumoto, K, Ye, M, Inoue, T, Kataoka, T. | | Deposit date: | 2009-11-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural basis for conformational dynamics of GTP-bound Ras protein
J.Biol.Chem., 285, 2010
|
|
