2ZK7
 
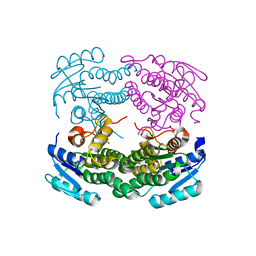 | | Structure of a C-terminal deletion mutant of Thermoplasma acidophilum aldohexose dehydrogenase (AldT) | | 分子名称: | Glucose 1-dehydrogenase related protein | | 著者 | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, N, Tamura, T. | | 登録日 | 2008-03-12 | | 公開日 | 2009-01-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | C-terminal tail derived from the neighboring subunit is critical for the activity of Thermoplasma acidophilum D-aldohexose dehydrogenase
Proteins, 74, 2009
|
|
1WMN
 
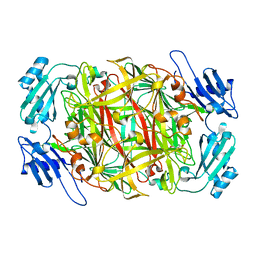 | | Crystal structure of topaquinone-containing amine oxidase activated by cobalt ion | | 分子名称: | COBALT (II) ION, Phenylethylamine oxidase | | 著者 | Okajima, T, Kishishita, S, Chiu, Y.C, Murakawa, T, Kim, M, Yamaguchi, H, Hirota, S, Kuroda, S, Tanizawa, K. | | 登録日 | 2004-07-13 | | 公開日 | 2005-08-02 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Reinvestigation of metal ion specificity for quinone cofactor biogenesis in bacterial copper amine oxidase
Biochemistry, 44, 2005
|
|
1WMO
 
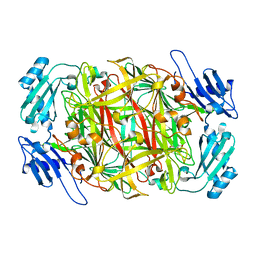 | | Crystal structure of topaquinone-containing amine oxidase activated by nickel ion | | 分子名称: | NICKEL (II) ION, Phenylethylamine oxidase | | 著者 | Okajima, T, Kishishita, S, Chiu, Y.C, Murakawa, T, Kim, M, Yamaguchi, H, Hirota, S, Kuroda, S, Tanizawa, K. | | 登録日 | 2004-07-13 | | 公開日 | 2005-08-02 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Reinvestigation of metal ion specificity for quinone cofactor biogenesis in bacterial copper amine oxidase
Biochemistry, 44, 2005
|
|
1WMP
 
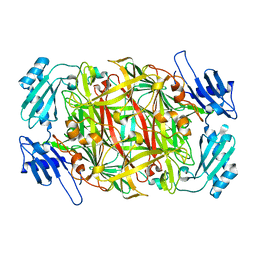 | | Crystal structure of amine oxidase complexed with cobalt ion | | 分子名称: | COBALT (II) ION, Phenylethylamine oxidase | | 著者 | Okajima, T, Kishishita, S, Chiu, Y.C, Murakawa, T, Kim, M, Yamaguchi, H, Hirota, S, Kuroda, S, Tanizawa, K. | | 登録日 | 2004-07-13 | | 公開日 | 2005-08-02 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Reinvestigation of metal ion specificity for quinone cofactor biogenesis in bacterial copper amine oxidase
Biochemistry, 44, 2005
|
|
2D1V
 
 | | Crystal structure of DNA-binding domain of Bacillus subtilis YycF | | 分子名称: | Transcriptional regulatory protein yycF | | 著者 | Okajima, T, Okada, A, Watanabe, T, Yamamoto, K, Tanizawa, K, Utsumi, R. | | 登録日 | 2005-09-01 | | 公開日 | 2006-09-12 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Response regulator YycF essential for bacterial growth: X-ray crystal structure of the DNA-binding domain and its PhoB-like DNA recognition motif
Febs Lett., 582, 2008
|
|
3AUS
 
 | |
3AUT
 
 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 in complex with NADH | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glucose 1-dehydrogenase 4 | | 著者 | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | 登録日 | 2011-02-16 | | 公開日 | 2012-02-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
3AY6
 
 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 A258F mutant in complex with NADH and D-glucose | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, Glucose 1-dehydrogenase 4, ... | | 著者 | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | 登録日 | 2011-04-29 | | 公開日 | 2012-05-23 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
3AY7
 
 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 G259A mutant | | 分子名称: | CHLORIDE ION, Glucose 1-dehydrogenase 4 | | 著者 | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | 登録日 | 2011-04-29 | | 公開日 | 2012-05-23 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
3AUU
 
 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 in complex with D-glucose | | 分子名称: | Glucose 1-dehydrogenase 4, beta-D-glucopyranose | | 著者 | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | 登録日 | 2011-02-16 | | 公開日 | 2012-02-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
3WTM
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Nitromethylene Analogue of Imidacloprid | | 分子名称: | 2-chloro-5-{[(2E)-2-(nitromethylidene)imidazolidin-1-yl]methyl}pyridine, Acetylcholine-binding protein | | 著者 | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | 登録日 | 2014-04-11 | | 公開日 | 2015-02-04 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
3WTL
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine Binding Protein Complexed with Nitromethylene Analogue of Imidacloprid | | 分子名称: | 2-chloro-5-{[(2E)-2-(nitromethylidene)imidazolidin-1-yl]methyl}pyridine, Acetylcholine-binding protein | | 著者 | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | 登録日 | 2014-04-11 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
3WTJ
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine Binding Protein Complexed with Thiacloprid | | 分子名称: | Acetylcholine-binding protein, {(2Z)-3-[(6-chloropyridin-3-yl)methyl]-1,3-thiazolidin-2-ylidene}cyanamide | | 著者 | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | 登録日 | 2014-04-11 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
3WTO
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Desnitro-imidacloprid | | 分子名称: | (2Z)-1-[(6-chloropyridin-3-yl)methyl]imidazolidin-2-imine, Acetylcholine-binding protein | | 著者 | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | 登録日 | 2014-04-11 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
3WTI
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Clothianidin | | 分子名称: | 1-[(2-chloro-1,3-thiazol-5-yl)methyl]-3-methyl-2-nitroguanidine, Acetylcholine-binding protein | | 著者 | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | 登録日 | 2014-04-11 | | 公開日 | 2015-02-04 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
3WTK
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Thiacloprid | | 分子名称: | Acetylcholine-binding protein, {(2Z)-3-[(6-chloropyridin-3-yl)methyl]-1,3-thiazolidin-2-ylidene}cyanamide | | 著者 | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | 登録日 | 2014-04-11 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
3WTH
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Imidacloprid | | 分子名称: | (2E)-1-[(6-chloropyridin-3-yl)methyl]-N-nitroimidazolidin-2-imine, Acetylcholine-binding protein | | 著者 | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | 登録日 | 2014-04-11 | | 公開日 | 2015-02-04 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
3WTN
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine Binding Protein Complexed with Desnitro-imidacloprid | | 分子名称: | (2Z)-1-[(6-chloropyridin-3-yl)methyl]imidazolidin-2-imine, Acetylcholine-binding protein, CADMIUM ION, ... | | 著者 | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | 登録日 | 2014-04-11 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
6JI6
 
 | |
3VRM
 
 | | Structure of cytochrome P450 Vdh mutant T107A with bound vitamin D3 | | 分子名称: | (1S,3Z)-3-[(2E)-2-[(1R,3AR,7AS)-7A-METHYL-1-[(2R)-6-METHYLHEPTAN-2-YL]-2,3,3A,5,6,7-HEXAHYDRO-1H-INDEN-4-YLIDENE]ETHYLI DENE]-4-METHYLIDENE-CYCLOHEXAN-1-OL, PROTOPORPHYRIN IX CONTAINING FE, Vitamin D(3) 25-hydroxylase | | 著者 | Nishioka, T, Yasutake, Y, Tamura, T. | | 登録日 | 2012-04-12 | | 公開日 | 2013-04-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | A single mutation at the ferredoxin binding site of p450 vdh enables efficient biocatalytic production of 25-hydroxyvitamin d3.
Chembiochem, 14, 2013
|
|
2ZXJ
 
 | |
1U19
 
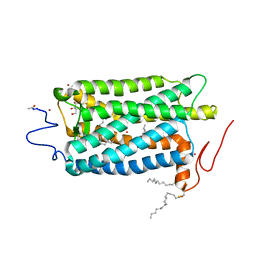 | | Crystal Structure of Bovine Rhodopsin at 2.2 Angstroms Resolution | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | 著者 | Okada, T, Sugihara, M, Bondar, A.N, Elstner, M, Entel, P, Buss, V. | | 登録日 | 2004-07-15 | | 公開日 | 2004-10-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure
J.Mol.Biol., 342, 2004
|
|
2E0Y
 
 | |
2E0X
 
 | |
2E0W
 
 | |
