4DWQ
 
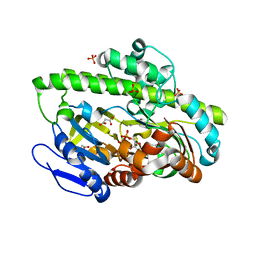 | | RNA ligase RtcB-GMP/Mn(2+) complex | | Descriptor: | GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, MALONATE ION, ... | | Authors: | Okada, C, Xia, S, Englert, M, Yao, M, Soll, D, Wang, J. | | Deposit date: | 2012-02-26 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3'-terminal phosphate and 5'-OH.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5B4D
 
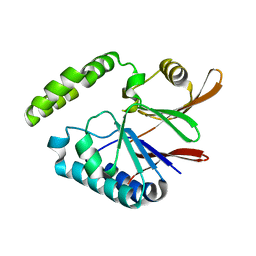 | | Crystal structure of H10N mutant of LpxH | | Descriptor: | GLYCEROL, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Okada, C, Wakabayashi, H, Yao, M, Tanaka, I. | | Deposit date: | 2016-04-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of the UDP-diacylglucosamine pyrophosphohydrase LpxH from Pseudomonas aeruginosa
Sci Rep, 6, 2016
|
|
5B4C
 
 | | Crystal structure of H10N mutant of LpxH with manganese | | Descriptor: | GLYCEROL, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Okada, C, Wakabayashi, H, Yao, M, Tanaka, I. | | Deposit date: | 2016-04-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structures of the UDP-diacylglucosamine pyrophosphohydrase LpxH from Pseudomonas aeruginosa
Sci Rep, 6, 2016
|
|
5B4A
 
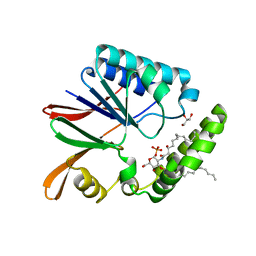 | | Crystal structure of LpxH with lipid X in spacegroup P21 | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, GLYCEROL, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Okada, C, Wakabayashi, H, Yao, M, Tanaka, I. | | Deposit date: | 2016-04-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of the UDP-diacylglucosamine pyrophosphohydrase LpxH from Pseudomonas aeruginosa
Sci Rep, 6, 2016
|
|
5B49
 
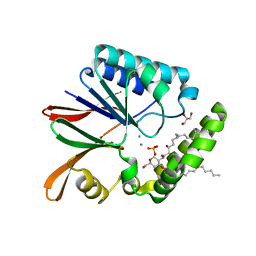 | | Crystal structure of LpxH with manganese from Pseudomonas aeruginosa | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Okada, C, Wakabayashi, H, Yao, M, Tanaka, I. | | Deposit date: | 2016-04-03 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of the UDP-diacylglucosamine pyrophosphohydrase LpxH from Pseudomonas aeruginosa
Sci Rep, 6, 2016
|
|
5B4B
 
 | | Crystal structure of LpxH with lipid X in spacegroup C2 | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, GLYCEROL, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Okada, C, Wakabayashi, H, Yao, M, Tanaka, I. | | Deposit date: | 2016-04-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of the UDP-diacylglucosamine pyrophosphohydrase LpxH from Pseudomonas aeruginosa
Sci Rep, 6, 2016
|
|
1UC2
 
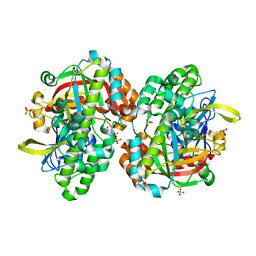 | | Hypothetical Extein Protein of PH1602 from Pyrococcus horikoshii | | Descriptor: | SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, hypothetical protein PH1602 | | Authors: | Okada, C, Maegawa, Y, Yao, M, Tanaka, I. | | Deposit date: | 2003-04-08 | | Release date: | 2004-05-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of an RtcB homolog protein (PH1602-extein protein) from Pyrococcus horikoshii reveals a novel fold
Proteins, 63, 2006
|
|
3A6P
 
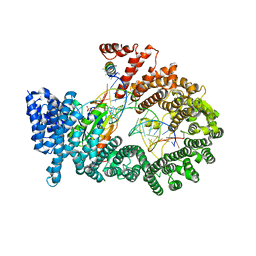 | | Crystal structure of Exportin-5:RanGTP:pre-miRNA complex | | Descriptor: | 13-mer peptide, Exportin-5, GTP-binding nuclear protein Ran, ... | | Authors: | Okada, C, Yamashita, E, Lee, S.J, Shibata, S, Katahira, J, Nakagawa, A, Yoneda, Y, Tsukihara, T. | | Deposit date: | 2009-09-07 | | Release date: | 2009-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | A high-resolution structure of the pre-microRNA nuclear export machinery
Science, 326, 2009
|
|
7XJV
 
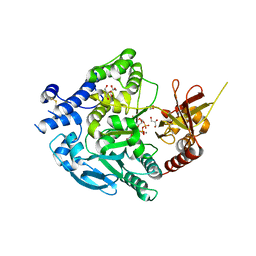 | | Crystal Structure of Alpha-1,3-mannosyltransferase MNT2 from Saccharomyces cerevisiae, Mn/GDP-mannose form | | Descriptor: | Alpha-1,3-mannosyltransferase MNT2, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | Authors: | Hira, D, Kadooka, C, Oka, T. | | Deposit date: | 2022-04-18 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Alpha-1,3-mannosyltransferase MNT2 from Saccharomyces cerevisiae, Mn/GDP-mannose form
To Be Published
|
|
1IY3
 
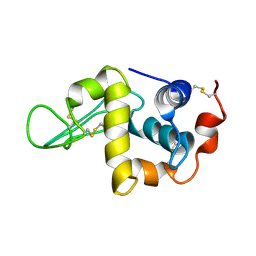 | | Solution Structure of the Human lysozyme at 4 degree C | | Descriptor: | Lysozyme | | Authors: | Kumeta, H, Miura, A, Kobashigawa, Y, Miura, K, Oka, C, Nitta, K, Nemoto, N, Tsuda, S. | | Deposit date: | 2002-07-15 | | Release date: | 2002-07-31 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in human lysozyme elucidated by three-dimensional NMR spectroscopy
Biochemistry, 42, 2003
|
|
1IY4
 
 | | Solution structure of the human lysozyme at 35 degree C | | Descriptor: | Lysozyme | | Authors: | Kumeta, H, Miura, A, Kobashigawa, Y, Miura, K, Oka, C, Nitta, K, Nemoto, N, Tsuda, S. | | Deposit date: | 2002-07-15 | | Release date: | 2002-07-31 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in human lysozyme elucidated by three-dimensional NMR spectroscopy
Biochemistry, 42, 2003
|
|
5KK2
 
 | | Architecture of fully occupied GluA2 AMPA receptor - TARP complex elucidated by single particle cryo-electron microscopy | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Zhao, Y, Chen, S, Yoshioka, C, Baconguis, I, Gouaux, E. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Architecture of fully occupied GluA2 AMPA receptor-TARP complex elucidated by cryo-EM.
Nature, 536, 2016
|
|
6NZ0
 
 | | Cryo-EM structure of AAV-2 in complex with AAVR PKD domains 1 and 2 | | Descriptor: | Capsid protein VP1, Dyslexia-associated protein KIAA0319-like protein, MAGNESIUM ION | | Authors: | Meyer, N.L, Xie, Q, Davulcu, O, Yoshioka, C, Chapman, M.S. | | Deposit date: | 2019-02-12 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure of the gene therapy vector, adeno-associated virus with its cell receptor, AAVR.
Elife, 8, 2019
|
|
6PZB
 
 | |
6PZI
 
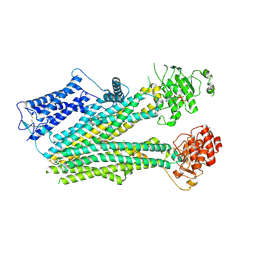 | | Cryo-EM structure of the pancreatic beta-cell SUR1 bound to ATP only | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8 | | Authors: | Shyng, S.L, Yoshioka, C, Martin, G.M, Sung, M.W. | | Deposit date: | 2019-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Mechanism of pharmacochaperoning in a mammalian K ATP channel revealed by cryo-EM.
Elife, 8, 2019
|
|
6PZC
 
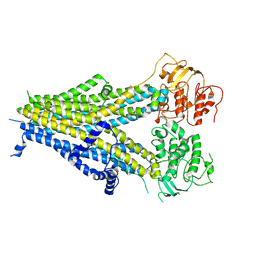 | |
7KFR
 
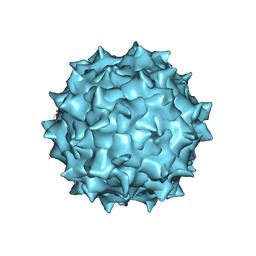 | |
6BQN
 
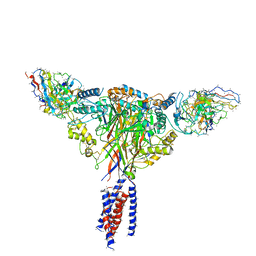 | | Cryo-EM structure of ENaC | | Descriptor: | 10D4 fab, 7B1 fab, EGFP-SCNN1G chimera, ... | | Authors: | Noreng, S, Bharadwaj, A, Posert, R, Yoshioka, C, Baconguis, I. | | Deposit date: | 2017-11-28 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the human epithelial sodium channel by cryo-electron microscopy.
Elife, 7, 2018
|
|
6PZA
 
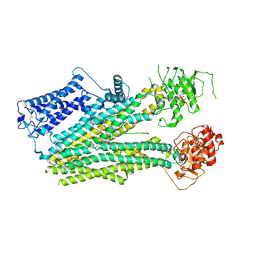 | | Cryo-EM structure of the pancreatic beta-cell SUR1 bound to ATP and glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Shyng, S.L, Yoshioka, C, Martin, G.M, Sung, M.W. | | Deposit date: | 2019-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Mechanism of pharmacochaperoning in a mammalian K ATP channel revealed by cryo-EM.
Elife, 8, 2019
|
|
6PZ9
 
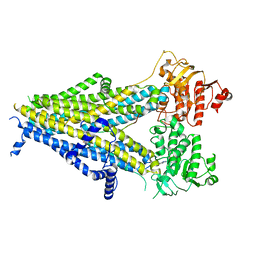 | | Cryo-EM structure of the pancreatic beta-cell SUR1 bound to ATP and repaglinide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ATP-sensitive inward rectifier potassium channel 11, ... | | Authors: | Shyng, S.L, Yoshioka, C, Martin, G.M, Sung, M.W. | | Deposit date: | 2019-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Mechanism of pharmacochaperoning in a mammalian K ATP channel revealed by cryo-EM.
Elife, 8, 2019
|
|
966C
 
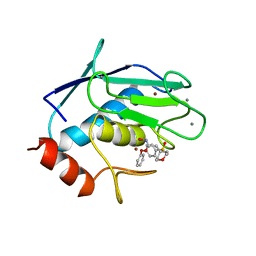 | | CRYSTAL STRUCTURE OF FIBROBLAST COLLAGENASE-1 COMPLEXED TO A DIPHENYL-ETHER SULPHONE BASED HYDROXAMIC ACID | | Descriptor: | CALCIUM ION, MMP-1, N-HYDROXY-2-[4-(4-PHENOXY-BENZENESULFONYL)-TETRAHYDRO-PYRAN-4-YL]-ACETAMIDE, ... | | Authors: | Lovejoy, B, Welch, A, Carr, S, Luong, C, Broka, C, Hendricks, R.T, Campbell, J, Walker, K, Martin, R, Van Wart, H, Browner, M.F. | | Deposit date: | 1998-08-07 | | Release date: | 1999-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of MMP-1 and -13 reveal the structural basis for selectivity of collagenase inhibitors.
Nat.Struct.Biol., 6, 1999
|
|
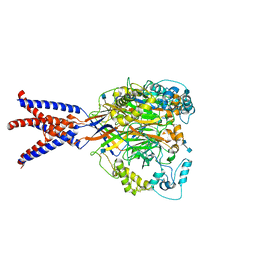
6AVE
 
 | |
4XZY
 
 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) from Porphyromonas gingivalis | | Descriptor: | GLYCEROL, Peptidase S46 | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2015-02-05 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
4Y01
 
 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) from Porphyromonas gingivalis | | Descriptor: | GLYCEROL, Peptidase S46 | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2015-02-05 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
4Y06
 
 | | Crystal structure of the DAP BII (G675R) dipeptide complex | | Descriptor: | Dipeptidyl aminopeptidase BII, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Sakamoto, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2015-02-05 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
