9LR5
 
 | |
5YLG
 
 | | Crystal Structure of LysM domain from pteris ryukyuensis chitinase A | | Descriptor: | 1,2-ETHANEDIOL, Chitinase A, ZINC ION | | Authors: | Ohnuma, T, Kitaoku, Y, Umemoto, N, Numata, T, Fukamizo, T. | | Deposit date: | 2017-10-17 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of LysM domain from pteris ryukyuensis chitinase A
To Be Published
|
|
8I5K
 
 | |
8I5J
 
 | |
7F88
 
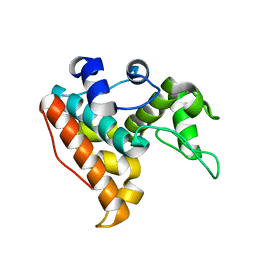 | |
4PXV
 
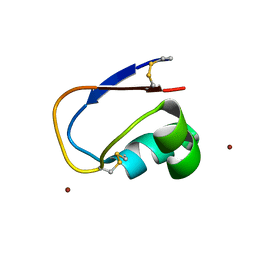 | |
5K2L
 
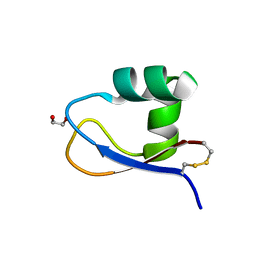 | | Crystal structure of LysM domain from Volvox carteri chitinase | | Descriptor: | 1,2-ETHANEDIOL, Chitinase, lysozyme | | Authors: | Kitaoku, Y, Numata, T, Fukamizo, T, Ohnuma, T. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Chitin oligosaccharide binding to the lysin motif of a novel type of chitinase from the multicellular green alga, Volvox carteri.
Plant Mol. Biol., 93, 2017
|
|
6JMS
 
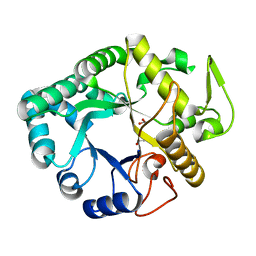 | | CJP38, a beta-1,3-glucanase and allergen of Cryptomeria japonica pollen | | Descriptor: | 1,2-ETHANEDIOL, Pollen allergen CJP38 | | Authors: | Takashima, T, Numata, T, Fukamizo, T, Ohnuma, T. | | Deposit date: | 2019-03-13 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CJP38, a beta-1,3-glucanase and allergen of Cryptomeria japonica pollen
To Be Published
|
|
4ZXE
 
 | | X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. | | Descriptor: | 1,2-ETHANEDIOL, Glucanase/Chitosanase, SULFATE ION | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-20 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
5BUM
 
 | | Crystal Structure of LysM domain from Equisetum arvense chitinase A | | Descriptor: | Chitinase A, SULFATE ION | | Authors: | Kitaoku, Y, Numata, T, Ohnuma, T, Taira, T, Fukamizo, T. | | Deposit date: | 2015-06-04 | | Release date: | 2016-06-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure, mechanism, and phylogeny of LysM-chitinase conjugates specifically found in fern plants.
Plant Sci., 321, 2022
|
|
5H7T
 
 | |
4ZY9
 
 | | X-ray crystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 | | Descriptor: | Glucanase/chitosanase | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-21 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
4ZZ8
 
 | | X-ray crystal structure of chitosan-binding module 2 in complex with chitotriose derived from chitosanase/glucanase from Paenibacillus sp. IK-5 | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Glucanase/chitosanase, ... | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
4ZZ5
 
 | | X-ray crystal structure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 | | Descriptor: | 1,2-ETHANEDIOL, Glucanase/chitosanase, SULFATE ION | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
5YZ6
 
 | | Solution structure of LysM domain from a chitinase derived from Volvox carteri | | Descriptor: | Chitinase, lysozyme | | Authors: | Kitaoku, Y, Nishimura, S, Fukamizo, T, Ohnuma, T. | | Deposit date: | 2017-12-13 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structures and chitin-binding properties of two N-terminal lysin motifs (LysMs) found in a chitinase from Volvox carteri.
Glycobiology, 29, 2019
|
|
5YZK
 
 | | Solution structure of LysM domain from a chitinase derived from Volvox carteri | | Descriptor: | Chitinase, lysozyme | | Authors: | Kitaoku, Y, Nishimura, S, Fukamizo, T, Ohnuma, T. | | Deposit date: | 2017-12-15 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structures and chitin-binding properties of two N-terminal lysin motifs (LysMs) found in a chitinase from Volvox carteri.
Glycobiology, 29, 2019
|
|
4DYG
 
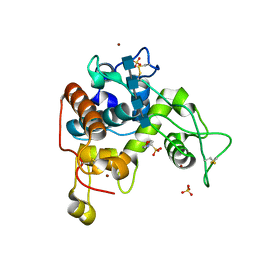 | | Crystal Structure of a Family GH-19 Chitinase from rye seeds in complex with (GlcNAc)4 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Basic endochitinase C, ... | | Authors: | Numata, T, Umemoto, N, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2012-02-29 | | Release date: | 2012-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and chitin oligosaccharide-binding mode of a 'loopful' family GH19 chitinase from rye, Secale cereale, seeds
Febs J., 279, 2012
|
|
4DWX
 
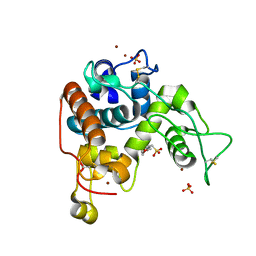 | | Crystal Structure of a Family GH-19 Chitinase from rye seeds | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Basic endochitinase C, SULFATE ION, ... | | Authors: | Numata, T, Umemoto, N, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2012-02-27 | | Release date: | 2012-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and chitin oligosaccharide-binding mode of a 'loopful' family GH19 chitinase from rye, Secale cereale, seeds
Febs J., 279, 2012
|
|
3WH1
 
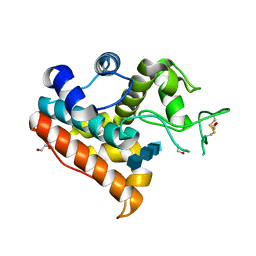 | | Crystal Structure of a Family GH19 Chitinase from Bryum coronatum in complex with (GlcNAc)4 at 1.0 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A | | Authors: | Numata, T, Umemoto, N, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2013-08-21 | | Release date: | 2014-03-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structure of a "loopless" GH19 chitinase in complex with chitin tetrasaccharide spanning the catalytic center.
Biochim.Biophys.Acta, 1844, 2014
|
|
3WIJ
 
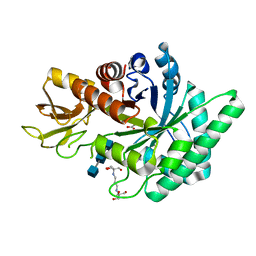 | | Crystal structure of a plant class V chitinase mutant from Cycas revoluta in complex with (GlcNAc)3 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Numata, T, Osawa, T, Umemoto, N, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2013-09-13 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a plant class V chitinase mutant from Cycas revoluta in complex with (GlcNAc)3
To be Published
|
|
4R5E
 
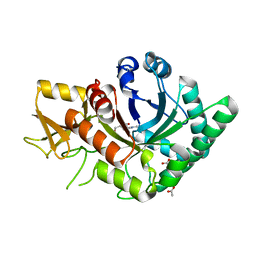 | | Crystal Structure of Family GH18 Chitinase from Cycas revoluta a Complex with Allosamidin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ACETATE ION, ALLOSAMIZOLINE, ... | | Authors: | Umemoto, N, Numata, T, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2014-08-21 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures and inhibitor binding properties of plant class V chitinases: the cycad enzyme exhibits unique structural and functional features.
Plant J., 82, 2015
|
|
4RL3
 
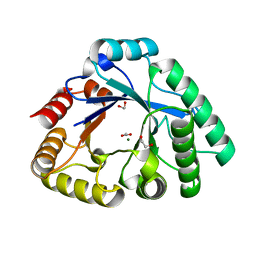 | | Crystal Structure of the Catalytic Domain of a family GH18 Chitinase from fern, Peteris ryukyuensis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Chitinase A, ... | | Authors: | Umemoto, N, Numata, T, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2014-10-15 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A class III chitinase without disulfide bonds from the fern, Pteris ryukyuensis: crystal structure and ligand-binding studies.
Planta, 242, 2015
|
|
4MNJ
 
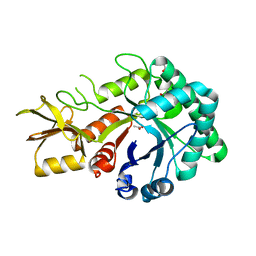 | | Crystal Structure of GH18 Chitinase from Cycad, Cycas revoluta | | Descriptor: | ACETATE ION, Chitinase A | | Authors: | Umemoto, N, Numata, T, Ohnuma, T, Osawa, T, Taira, T, Fukamizo, T. | | Deposit date: | 2013-09-11 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of GH18 Chitinase from Cycad, Cycas revoluta
To be Published
|
|
4MNL
 
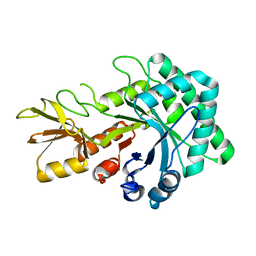 | | Crystal Structure of GH18 Chitinase (G77W/E119Q mutant) from Cycas revoluta in complex with (GlcNAc)4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Chitinase A | | Authors: | Umemoto, N, Numata, T, Ohnuma, T, Osawa, T, Taira, T, Fukamizo, T. | | Deposit date: | 2013-09-11 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of GH18 Chitinase form Cycad, Cycas revoluta
To be Published
|
|
4MNK
 
 | | Crystal Structure of GH18 Chitinase from Cycas revoluta in complex with (GlcNAc)3 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Umemoto, N, Numata, T, Ohnuma, T, Osawa, T, Taira, T, Fukamizo, T. | | Deposit date: | 2013-09-11 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystal Structure of GH18 Chitinase from Cycad, Cycas revoluta
To be Published
|
|
