6ZRT
 
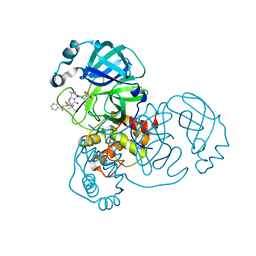 | | Crystal structure of SARS CoV2 main protease in complex with inhibitor Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, DIMETHYL SULFOXIDE, Main Protease | | Authors: | Oerlemans, R, Wang, W, Lunev, S, Domling, A, Groves, M.R. | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Repurposing the HCV NS3-4A protease drug boceprevir as COVID-19 therapeutics.
Rsc Med Chem, 12, 2020
|
|
6ZRU
 
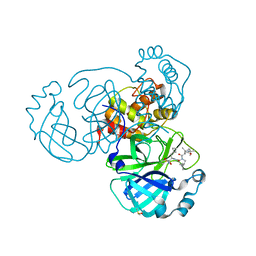 | | Crystal structure of SARS CoV2 main protease in complex with inhibitor Boceprevir | | Descriptor: | DIMETHYL SULFOXIDE, Main Protease, boceprevir (bound form) | | Authors: | Oerlemans, R, Wang, W, Lunev, S, Domling, A, Groves, M.R. | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Repurposing the HCV NS3-4A protease drug boceprevir as COVID-19 therapeutics.
Rsc Med Chem, 12, 2020
|
|
7NT2
 
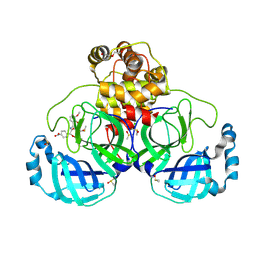 | | Crystal structure of SARS CoV2 main protease in complex with FSP006 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.145 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NT1
 
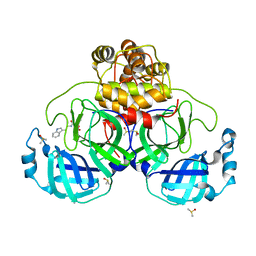 | | Crystal structure of SARS CoV2 main protease in complex with FSP007 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, [(2R)-1-[2-(1H-indol-3-yl)ethylamino]-1-oxidanylidene-butan-2-yl] prop-2-enoate | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NT3
 
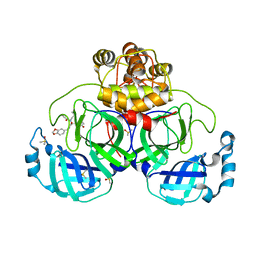 | | Crystal structure of SARS CoV2 main protease in complex with FSCU015 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(1~{S})-2-(1,3-benzodioxol-5-ylmethylamino)-1-(3-hydroxyphenyl)-2-oxidanylidene-ethyl]-~{N}-propyl-prop-2-enamide | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.325 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NTV
 
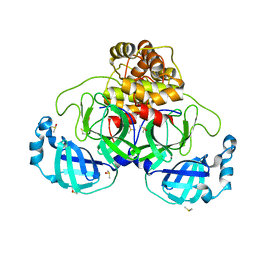 | | Crystal structure of SARS CoV2 main protease in complex with DN_EG_002 (modelled using PanDDA event map) | | Descriptor: | 2-acetamido-N-cyclopropyl-5-phenyl-thiophene-3-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-10 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.065 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NUK
 
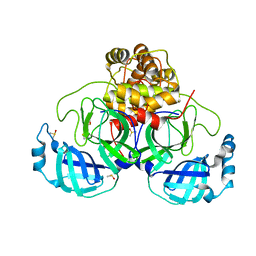 | | Crystal structure of SARS CoV2 main protease in complex with EG009 (modelled using PanDDA event map) | | Descriptor: | 2-[2-chloranylethanoyl(propyl)amino]-~{N}-(2-methoxyphenyl)ethanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-12 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8OGH
 
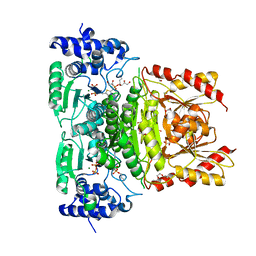 | | Truncated 1-deoxy-D-xylulose 5-phosphate synthase (DXPS) from Mycobacterium tuberculosis with butylacetylphosphonate (BAP) bound | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose-5-phosphate synthase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gawriljuk, O.V, Oerlemans, R, Groves, R.M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Mycobacterium tuberculosis 1-Deoxy-D-Xylulose 5-Phosphate Synthase in Complex with Butylacetylphosphonate
Crystals, 13, 2023
|
|
8R2H
 
 | | Cryo-EM structure of 1-deoxy-D-xylulose 5-phosphate synthase (DXPS) from Plasmodium falciparum | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Gawriljuk, V.O, Godoy, A.S, Oerlemans, R, Groves, M.R. | | Deposit date: | 2023-11-06 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Cryo-EM structure of 1-deoxy-D-xylulose 5-phosphate synthase DXPS from Plasmodium falciparum reveals a distinct N-terminal domain.
Nat Commun, 15, 2024
|
|
