2AHJ
 
 | | NITRILE HYDRATASE COMPLEXED WITH NITRIC OXIDE | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, FE (III) ION, NITRIC OXIDE, ... | | Authors: | Nagashima, S, Nakasako, M, Dohmae, N, Tsujimura, M, Takio, K, Odaka, M, Yohda, M, Kamiya, N, Endo, I. | | Deposit date: | 1997-12-24 | | Release date: | 1999-01-27 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel non-heme iron center of nitrile hydratase with a claw setting of oxygen atoms.
Nat.Struct.Biol., 5, 1998
|
|
6L98
 
 | | Crystalline cast nephropathy-causing Bence-Jones protein AK: An entire immunoglobulin lambda light chain dimer | | Descriptor: | Bence-Jones protein lambda light chain AK | | Authors: | Nakagaki, T, Noguchi, K, Yohda, M, Odaka, M, Wakui, H, Matsumura, H. | | Deposit date: | 2019-11-08 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Multiple Myeloma-Associated Ig Light Chain Crystalline Cast Nephropathy.
Kidney Int Rep, 5, 2020
|
|
8IMR
 
 | | Structure of ligand-free human macrophage migration inhibitory factor | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, ... | | Authors: | Sugishima, K, Noguchi, K, Yohda, M, Odaka, M, Matsumura, H. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Identification of methotrexate as an inhibitor of macrophage migration inhibitory factor by high-resolution crystal structure analysis
To Be Published
|
|
1HSQ
 
 | | SOLUTION STRUCTURE OF THE SH3 DOMAIN OF PHOSPHOLIPASE CGAMMA | | Descriptor: | PHOSPHOLIPASE C-GAMMA (SH3 DOMAIN) | | Authors: | Kohda, D, Hatanaka, H, Odaka, M, Inagaki, F. | | Deposit date: | 1994-06-13 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of phospholipase C-gamma.
Cell(Cambridge,Mass.), 72, 1993
|
|
2HSP
 
 | | SOLUTION STRUCTURE OF THE SH3 DOMAIN OF PHOSPHOLIPASE CGAMMA | | Descriptor: | PHOSPHOLIPASE C-GAMMA (SH3 DOMAIN) | | Authors: | Kohda, D, Hatanaka, H, Odaka, M, Inagaki, F. | | Deposit date: | 1994-06-13 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of phospholipase C-gamma.
Cell(Cambridge,Mass.), 72, 1993
|
|
7XTX
 
 | | High resolution crystal structure of human macrophage migration inhibitory factor in complex with methotrexate | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, ... | | Authors: | Sugishima, K, Noguchi, K, Yohda, M, Odaka, M, Matsumura, H. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Identification of methotrexate as an inhibitor of macrophage migration inhibitory factor by high-resolution crystal structure analysis
To Be Published
|
|
7XVX
 
 | |
5B19
 
 | | Picrophilus torridus aspartate racemase | | Descriptor: | Aspartate racemase, L(+)-TARTARIC ACID | | Authors: | Aihara, T, Ito, T, Yamanaka, Y, Noguchi, K, Odaka, M, Sekine, M, Homma, H, Yohda, M. | | Deposit date: | 2015-11-30 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural and functional characterization of aspartate racemase from the acidothermophilic archaeon Picrophilus torridus
Extremophiles, 20, 2016
|
|
4Z9F
 
 | | Halohydrin hydrogen-halide-lyase, HheA | | Descriptor: | CHLORIDE ION, Halohydrin epoxidase A | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2015-04-10 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of halohydrin hydrogen-halide-lyases from Corynebacterium sp. N-1074
Proteins, 83, 2015
|
|
4ZU3
 
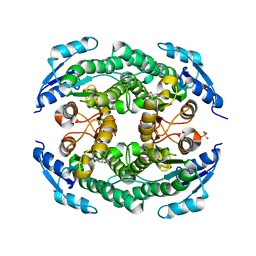 | | Halohydrin hydrogen-halide-lyases, HheB | | Descriptor: | 3-hydroxypentanedinitrile, Halohydrin epoxidase B, MAGNESIUM ION, ... | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2015-05-15 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of halohydrin hydrogen-halide-lyases from Corynebacterium sp. N-1074
Proteins, 83, 2015
|
|
4ZD6
 
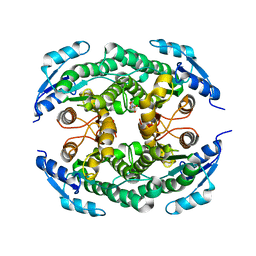 | | Halohydrin hydrogen-halide-lyase, HheB | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Halohydrin epoxidase B, ... | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2015-04-17 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of halohydrin hydrogen-halide-lyases from Corynebacterium sp. N-1074
Proteins, 83, 2015
|
|
5B12
 
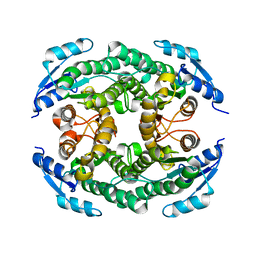 | | Crystal structure of the B-type halohydrin hydrogen-halide-lyase mutant F71W/Q125T/D199H from Corynebacterium sp. N-1074 | | Descriptor: | CHLORIDE ION, Halohydrin epoxidase B | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Odaka, M, Yohda, M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.721 Å) | | Cite: | Improvement of enantioselectivity of the B-type halohydrin hydrogen-halide-lyase from Corynebacterium sp. N-1074
J.Biosci.Bioeng., 122, 2016
|
|
2DD5
 
 | | Thiocyanate hydrolase (SCNase) from Thiobacillus thioparus native holo-enzyme | | Descriptor: | COBALT (III) ION, SULFATE ION, Thiocyanate hydrolase alpha subunit, ... | | Authors: | Arakawa, T, Kawano, Y, Kataoka, S, Katayama, Y, Kamiya, N, Yohda, M, Odaka, M. | | Deposit date: | 2006-01-19 | | Release date: | 2007-01-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of thiocyanate hydrolase: a new nitrile hydratase family protein with a novel five-coordinate cobalt(III) center.
J.Mol.Biol., 366, 2007
|
|
2DD4
 
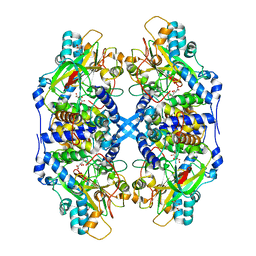 | | Thiocyanate hydrolase (SCNase) from Thiobacillus thioparus recombinant apo-enzyme | | Descriptor: | L(+)-TARTARIC ACID, Thiocyanate hydrolase alpha subunit, Thiocyanate hydrolase beta subunit, ... | | Authors: | Arakawa, T, Kawano, Y, Kataoka, S, Katayama, Y, Kamiya, N, Yohda, M, Odaka, M. | | Deposit date: | 2006-01-19 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of thiocyanate hydrolase: a new nitrile hydratase family protein with a novel five-coordinate cobalt(III) center.
J.Mol.Biol., 366, 2007
|
|
2DXC
 
 | | Recombinant thiocyanate hydrolase, fully-matured form | | Descriptor: | COBALT (III) ION, L(+)-TARTARIC ACID, Thiocyanate hydrolase subunit alpha, ... | | Authors: | Arakawa, T, Kawano, Y, Katayama, Y, Yohda, M, Odaka, M. | | Deposit date: | 2006-08-25 | | Release date: | 2007-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Catalytic Activation of Thiocyanate Hydrolase Involving Metal-Ligated Cysteine Modification
J.Am.Chem.Soc., 131, 2009
|
|
2DXB
 
 | | Recombinant thiocyanate hydrolase comprising partially-modified cobalt centers | | Descriptor: | COBALT (III) ION, PHOSPHATE ION, Thiocyanate hydrolase subunit alpha, ... | | Authors: | Arakawa, T, Kawano, Y, Katayama, Y, Yohda, M, Odaka, M. | | Deposit date: | 2006-08-25 | | Release date: | 2007-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis for Catalytic Activation of Thiocyanate Hydrolase Involving Metal-Ligated Cysteine Modification
J.Am.Chem.Soc., 131, 2009
|
|
2DX7
 
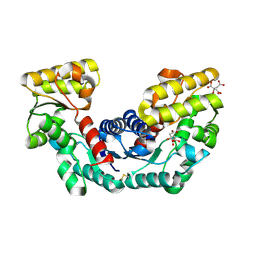 | | Crystal structure of Pyrococcus horikoshii OT3 aspartate racemase complex with citric acid | | Descriptor: | CITRIC ACID, aspartate racemase | | Authors: | Ohtaki, A, Arakawa, T, Iizuka, R, Odaka, M, Yohda, M. | | Deposit date: | 2006-08-24 | | Release date: | 2007-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of aspartate racemase complexed with a dual substrate analogue, citric acid, and implications for the reaction mechanism.
Proteins, 70, 2008
|
|
2CZ1
 
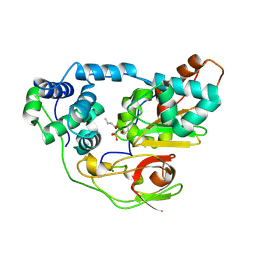 | | photo-activation state of Fe-type NHase with n-BA in anaerobic condition | | Descriptor: | BUTANOIC ACID, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Kawano, Y, Hashimoto, K, Odaka, M, Nakayama, H, Takio, K, Endo, I, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-09 | | Release date: | 2006-01-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | photo-activation state of Fe-type NHase with n-BA in anaerobic condition
To be Published
|
|
2CYZ
 
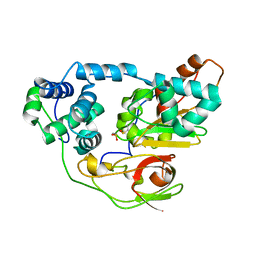 | | photo-activation state of Fe-type NHase in anaerobic condition | | Descriptor: | FE (III) ION, MAGNESIUM ION, Nitrile hydratase subunit alpha, ... | | Authors: | Kawano, Y, Hashimoto, K, Odaka, M, Nakayama, H, Takio, K, Endo, I, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-09 | | Release date: | 2006-01-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | photo-activation state of Fe-type NHase in anaerobic condition
To be Published
|
|
2CZ0
 
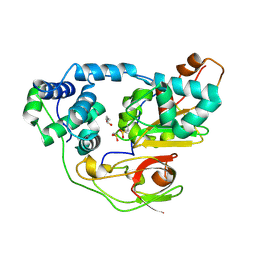 | | photo-activation state of Fe-type NHase in aerobic condition | | Descriptor: | BUTANOIC ACID, FE (III) ION, Nitrile hydratase subunit alpha, ... | | Authors: | Kawano, Y, Hashimoto, K, Odaka, M, Nakayama, H, Takio, K, Endo, I, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-09 | | Release date: | 2006-01-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | photo-activation state of Fe-type NHase in aerobic condition
To be Published
|
|
3VQJ
 
 | | Crystal Structutre of Thiobacillus thioparus THI115 Carbonyl Sulfide Hydrolase | | Descriptor: | Carbonyl sulfide hydrolase, SODIUM ION, ZINC ION | | Authors: | Katayama, Y, Noguchi, K, Ogawa, T, Ohtaki, A, Odaka, M, Yohda, M. | | Deposit date: | 2012-03-24 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Carbonyl Sulfide Hydrolase from Thiobacillus thioparus Strain THI115 Is One of the beta-Carbonic Anhydrase Family Enzymes
J.Am.Chem.Soc., 135, 2013
|
|
3VRK
 
 | | Crystal Structutre of Thiobacillus thioparus THI115 Carbonyl Sulfide Hydrolase / Thiocyanate complex | | Descriptor: | Carbonyl sulfide hydrolase, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Katayama, Y, Noguchi, K, Ogawa, T, Ohtaki, A, Odaka, M, Yohda, M. | | Deposit date: | 2012-04-11 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Carbonyl Sulfide Hydrolase from Thiobacillus thioparus Strain THI115 Is One of the beta-Carbonic Anhydrase Family Enzymes
J.Am.Chem.Soc., 135, 2013
|
|
3VYH
 
 | | Crystal structure of aW116R mutant of nitrile hydratase from Pseudonocardia thermophilla | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta | | Authors: | Yamanaka, Y, Sato, M, Arakawa, T, Namima, S, Hori, S, Ohtaki, A, Noguchi, K, Katayama, Y, Yohda, M, Odaka, M. | | Deposit date: | 2012-09-25 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Effects of argnine residue around the substrate pocket on the substrate specificity of thiocyanate hydrolase
To be published
|
|
3VYG
 
 | | Crystal structure of Thiocyanate hydrolase mutant R136W | | Descriptor: | COBALT (III) ION, L(+)-TARTARIC ACID, Thiocyanate hydrolase subunit alpha, ... | | Authors: | Yamanaka, Y, Sato, M, Arakawa, T, Namima, S, Hori, S, Ohtaki, A, Noguchi, K, Katayama, Y, Yohda, M, Odaka, M. | | Deposit date: | 2012-09-25 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Effects of argnine residue around the substrate pocket on the substrate specificity of thiocyanate hydrolase
To be published
|
|
3WVE
 
 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, before photo-activation | | Descriptor: | 2,2-dimethylpropanenitrile, CHLORIDE ION, FE (III) ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2014-05-17 | | Release date: | 2015-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
