6V24
 
 | |
3FIV
 
 | | CRYSTAL STRUCTURE OF FELINE IMMUNODEFICIENCY VIRUS PROTEASE COMPLEXED WITH A SUBSTRATE | | Descriptor: | ACE-ALN-VAL-LEU-ALA-GLU-ALN-NH2, FELINE IMMUNODEFICIENCY VIRUS PROTEASE, SULFATE ION | | Authors: | Schalk-Hihi, C, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1997-07-09 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the inactive D30N mutant of feline immunodeficiency virus protease complexed with a substrate and an inhibitor.
Biochemistry, 36, 1997
|
|
6FIV
 
 | | STRUCTURAL STUDIES OF HIV AND FIV PROTEASES COMPLEXED WITH AN EFFICIENT INHIBITOR OF FIV PR | | Descriptor: | RETROPEPSIN, SULFATE ION, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Li, M, Lee, T, Morris, G, Laco, G, Wong, C, Olson, A, Elder, J, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-12-02 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of FIV and HIV-1 proteases complexed with an efficient inhibitor of FIV protease
Proteins, 38, 2000
|
|
6D45
 
 | | L89S Mutant of FeBMb Sperm Whale Myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bhagi-Damodaran, A, Mirts, E.N, Sandoval, B, Lu, Y. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Heme redox potentials hold the key to reactivity differences between nitric oxide reductase and heme-copper oxidase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4YDX
 
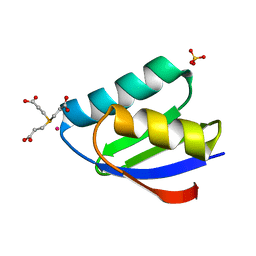 | | Crystal structure of cisplatin bound to a human copper chaperone (monomer) - new refinement | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, Copper transport protein ATOX1, PLATINUM (II) ION, ... | | Authors: | Shabalin, I.G, Boal, A.K, Dauter, Z, Jaskolski, M, Minor, W, Rosenzweig, A.C, Wlodawer, A. | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YEA
 
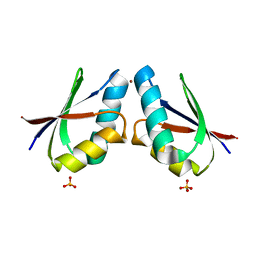 | | Crystal structure of cisplatin bound to a human copper chaperone (dimer) - new refinement | | Descriptor: | COPPER (II) ION, Copper transport protein ATOX1, SULFATE ION | | Authors: | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YEN
 
 | | Room temperature X-ray diffraction studies of cisplatin binding to HEWL in DMSO media after 14 months of crystal storage - new refinement | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YEM
 
 | | Carboplatin binding to HEWL in NaBr crystallisation conditions studied at an X-ray wavelength of 0.9163A - new refinement | | Descriptor: | ACETATE ION, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YEO
 
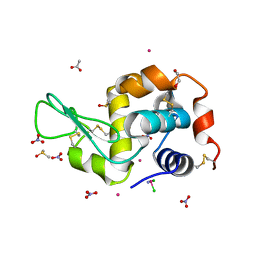 | | Triclinic HEWL co-crystallised with cisplatin, studied at a data collection temperature of 150K - new refinement | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cisplatin, ... | | Authors: | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8ECE
 
 | | E. coli L-asparaginase II mutant (V27T) in complex with L-Glu | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, L-asparaginase 2 | | Authors: | Strzelczyk, P, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The E. coli L-asparaginase V27T mutant: structural and functional characterization and comparison with theoretical predictions.
Febs Lett., 596, 2022
|
|
8ECD
 
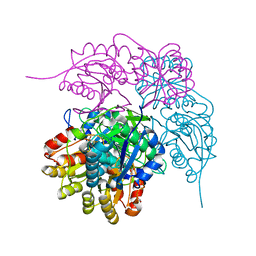 | | E. coli L-asparaginase II mutant (V27T) in complex with L-Asp | | Descriptor: | 1,2-ETHANEDIOL, ASPARTIC ACID, CITRIC ACID, ... | | Authors: | Strzelczyk, P, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The E. coli L-asparaginase V27T mutant: structural and functional characterization and comparison with theoretical predictions.
Febs Lett., 596, 2022
|
|
5VNU
 
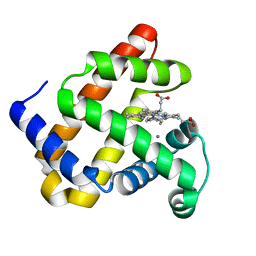 | | Nonheme Iron Replacement in a Biosynthetic Nitric Oxide Reductase Model Performing O2 Reduction to Water: Mn-bound FeBMb | | Descriptor: | MANGANESE (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Reed, J, Shi, Y, Zhu, Q, Chakraborty, S, Mirs, E.N, Petrik, I.D, Bhagi-Damodaran, A, Ross, M, Moenne-Loccoz, P, Zhang, Y, Lu, Y. | | Deposit date: | 2017-05-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.584 Å) | | Cite: | Manganese and Cobalt in the Nonheme-Metal-Binding Site of a Biosynthetic Model of Heme-Copper Oxidase Superfamily Confer Oxidase Activity through Redox-Inactive Mechanism.
J. Am. Chem. Soc., 139, 2017
|
|
5VRT
 
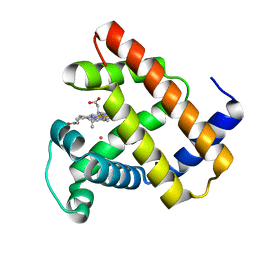 | | Nonheme Iron Replacement in a Biosynthetic Nitric Oxide Reductase Model Performing O2 Reduction to Water: Co-bound FeBMb | | Descriptor: | COBALT (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Reed, J, Shi, Y, Zhu, Q, Chakraborty, S, Mirs, E.N, Petrik, I.D, Bhagi-Damodaran, A, Ross, M, Moenne-Loccoz, P, Zhang, Y, Lu, Y. | | Deposit date: | 2017-05-11 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Manganese and Cobalt in the Nonheme-Metal-Binding Site of a Biosynthetic Model of Heme-Copper Oxidase Superfamily Confer Oxidase Activity through Redox-Inactive Mechanism.
J. Am. Chem. Soc., 139, 2017
|
|
5O9I
 
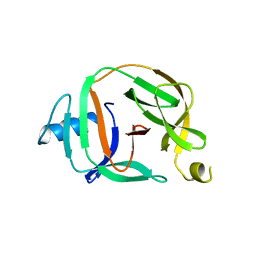 | | Crystal structure of transcription factor IIB Mvu mini-intein | | Descriptor: | Transcription initiation factor IIB,Transcription initiation factor IIB | | Authors: | Mikula, K.M, Iwai, H, Li, M, Wlodawer, A. | | Deposit date: | 2017-06-19 | | Release date: | 2017-11-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Persistence of Homing Endonucleases in Transcription Factor IIB Inteins.
J. Mol. Biol., 429, 2017
|
|
7BPF
 
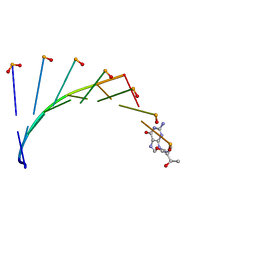 | | Structure of L-threoninol nucleic acid - RNA complex | | Descriptor: | L-aTNA (3'-(*GP*CP*AP*GP*CP*AP*GP*C)-1'), RNA (5'-R(*GP*CP*UP*GP*CP*(5BU)P*GP*C)-3') | | Authors: | Kamiya, Y, Satoh, T, Kodama, A, Suzuki, T, Uchiyama, S, Kato, K, Asanuma, H. | | Deposit date: | 2020-03-22 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of heteroduplexes of L-aTNA/RNA and SNA/RNA
Commun Chem, 2020
|
|
7BPG
 
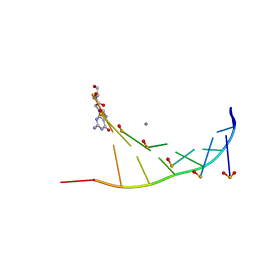 | | Structure of serinol nucleic acid - RNA complex | | Descriptor: | CALCIUM ION, RNA (5'-R(*GP*CP*UP*GP*CP*(5BU)P*GP*C)-3'), SNA (S-(F7R)(F7X)(F7O)(F7R)(F7X)(F7O)(F7R)(F7U)-R) | | Authors: | Kamiya, Y, Satoh, T, Kodama, A, Suzuki, T, Uchiyama, S, Kato, K, Asanuma, H. | | Deposit date: | 2020-03-22 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of heteroduplexes of L-aTNA/RNA and SNA/RNA
Commun Chem, 2020
|
|
6YI0
 
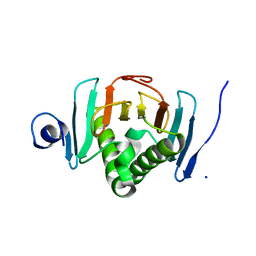 | | Human histidine triad nucleotide-binding protein 2 (hHINT2) refined to 1.65 A in P41212 space group | | Descriptor: | Histidine triad nucleotide-binding protein 2, mitochondrial, SODIUM ION | | Authors: | Dolot, R.D, Wlodarczyk, A, Bujacz, G.D, Nawrot, B.C. | | Deposit date: | 2020-03-31 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Biochemical, crystallographic and biophysical characterization of histidine triad nucleotide-binding protein 2 with different ligands including a non-hydrolyzable analog of Ap4A.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
6YPX
 
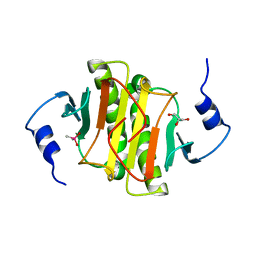 | | Human histidine triad nucleotide-binding protein 2 (hHINT2) refined to 2.11 A in C2221 space group | | Descriptor: | CACODYLATE ION, GLYCEROL, Histidine triad nucleotide-binding protein 2, ... | | Authors: | Dolot, R.D, Wlodarczyk, A, Bujacz, G.D, Nawrot, B.C. | | Deposit date: | 2020-04-16 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Biochemical, crystallographic and biophysical characterization of histidine triad nucleotide-binding protein 2 with different ligands including a non-hydrolyzable analog of Ap4A.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
6WYW
 
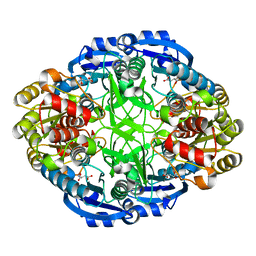 | | Crystal structure of Pseudomonas 7A Glutaminase-Asparaginase in complex with L-Asp at pH 4.5 | | Descriptor: | ASPARTIC ACID, Glutaminase-asparaginase | | Authors: | Strzelczyk, P, Zhang, D, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2020-05-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Generalized enzymatic mechanism of catalysis by tetrameric L-asparaginases from mesophilic bacteria.
Sci Rep, 10, 2020
|
|
6WYX
 
 | | Crystal structure of Pseudomonas 7A Glutaminase-Asparaginase in complex with L-Asp at pH 5.0 | | Descriptor: | ASPARTIC ACID, GLYCEROL, Glutaminase-asparaginase | | Authors: | Strzelczyk, P, Zhang, D, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2020-05-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Generalized enzymatic mechanism of catalysis by tetrameric L-asparaginases from mesophilic bacteria.
Sci Rep, 10, 2020
|
|
6WZ8
 
 | | Complex of Pseudomonas 7A Glutaminase-Asparaginase with Citrate Anion at pH 5.5. Covalent Acyl-Enzyme Intermediate | | Descriptor: | CITRIC ACID, Glutaminase-asparaginase | | Authors: | Strzelczyk, P, Zhang, D, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2020-05-13 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Generalized enzymatic mechanism of catalysis by tetrameric L-asparaginases from mesophilic bacteria.
Sci Rep, 10, 2020
|
|
6WZ4
 
 | | Complex of mutant (K173M) of Pseudomonas 7A Glutaminase-Asparaginase with L-Asp at pH 6. Covalent acyl-enzyme intermediate | | Descriptor: | ASPARTIC ACID, Glutaminase-asparaginase | | Authors: | Strzelczyk, P, Zhang, D, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2020-05-13 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Generalized enzymatic mechanism of catalysis by tetrameric L-asparaginases from mesophilic bacteria.
Sci Rep, 10, 2020
|
|
6YQD
 
 | | Human histidine triad nucleotide-binding protein 2 (hHINT2) refined to 1.41 A in P212121 space group | | Descriptor: | Histidine triad nucleotide-binding protein 2, mitochondrial, POTASSIUM ION | | Authors: | Dolot, R.D, Wlodarczyk, A, Bujacz, G.D, Nawrot, B.C. | | Deposit date: | 2020-04-16 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.407 Å) | | Cite: | Biochemical, crystallographic and biophysical characterization of histidine triad nucleotide-binding protein 2 with different ligands including a non-hydrolyzable analog of Ap4A.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
6WYZ
 
 | | Crystal structure of Pseudomonas 7A Glutaminase-Asparaginase (mutant K173M) in complex with D-Glu at pH 5.5 | | Descriptor: | D-GLUTAMIC ACID, Glutaminase-asparaginase | | Authors: | Strzelczyk, P, Zhang, D, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2020-05-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Generalized enzymatic mechanism of catalysis by tetrameric L-asparaginases from mesophilic bacteria.
Sci Rep, 10, 2020
|
|
6YPR
 
 | | Human histidine triad nucleotide-binding protein 2 (hHINT2) refined to 1.26 A in H32 space group | | Descriptor: | GLYCEROL, Histidine triad nucleotide-binding protein 2, mitochondrial | | Authors: | Dolot, R.D, Wlodarczyk, A, Bujacz, G.D, Nawrot, B.C. | | Deposit date: | 2020-04-16 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Biochemical, crystallographic and biophysical characterization of histidine triad nucleotide-binding protein 2 with different ligands including a non-hydrolyzable analog of Ap4A.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
