2AN7
 
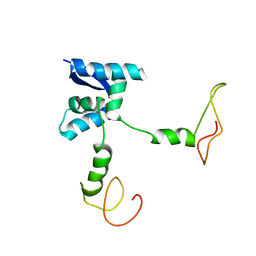 | | Solution structure of the bacterial antidote ParD | | Descriptor: | Protein parD | | Authors: | Oberer, M, Zangger, K, Gruber, K, Keller, W. | | Deposit date: | 2005-08-11 | | Release date: | 2006-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of ParD, the antidote of the ParDE toxin antitoxin module, provides the structural basis for DNA and toxin binding.
Protein Sci., 16, 2007
|
|
6EIC
 
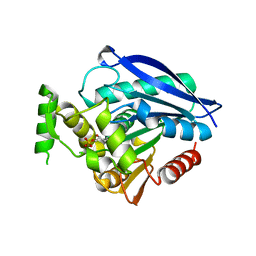 | | Crystal structure of Rv0183, a Monoglyceride Lipase from Mycobacterium Tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Mycobacterium Tuberculosis Monoglyceride Lipase, NITRATE ION, ... | | Authors: | Aschauer, P, Pavkov-Keller, T, Oberer, M. | | Deposit date: | 2017-09-19 | | Release date: | 2018-06-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of monoacylglycerol lipase from M. tuberculosis reveals the basis for specific inhibition.
Sci Rep, 8, 2018
|
|
2ADL
 
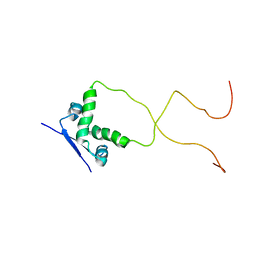 | | Solution structure of the bacterial antitoxin CcdA: Implications for DNA and toxin binding | | Descriptor: | CcdA | | Authors: | Madl, T, VanMelderen, L, Oberer, M, Keller, W, Khatai, L, Zangger, K. | | Deposit date: | 2005-07-20 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2ADN
 
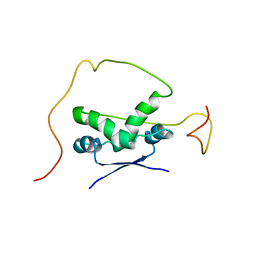 | | Solution structure of the bacterial antitoxin CcdA: Implications for DNA and toxin binding | | Descriptor: | CcdA | | Authors: | Madl, T, VanMelderen, L, Oberer, M, Keller, W, Khatai, L, Zangger, K. | | Deposit date: | 2005-07-20 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2LYC
 
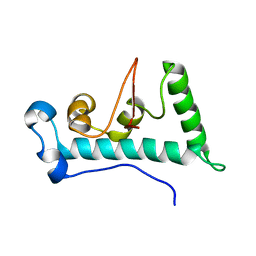 | | Structure of C-terminal domain of Ska1 | | Descriptor: | Spindle and kinetochore-associated protein 1 homolog | | Authors: | Boeszoermenyi, A, Schmidt, J.C, Markus, M, Oberer, M, Cheeseman, I.M, Wagner, G, Arthanari, H. | | Deposit date: | 2012-09-14 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The kinetochore-bound ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments.
Dev.Cell, 23, 2012
|
|
8AXC
 
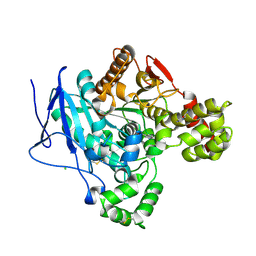 | | Crystal structure of mouse Ces2c | | Descriptor: | Acylcarnitine hydrolase, CHLORIDE ION, NICOTINAMIDE, ... | | Authors: | Eisner, H, Riegler-Berket, L, Rodriguez Gamez, C, Sagmeister, T, Chalhoub, G, Darnhofer, B, Panikkaveetil Jawaharlal, J, Birner-Gruenberger, R, Pavkov-Keller, T, Haemmerle, G, Schoiswohl, G, Oberer, M. | | Deposit date: | 2022-08-31 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The Crystal Structure of Mouse Ces2c, a Potential Ortholog of Human CES2, Shows Structural Similarities in Substrate Regulation and Product Release to Human CES1.
Int J Mol Sci, 23, 2022
|
|
7OZM
 
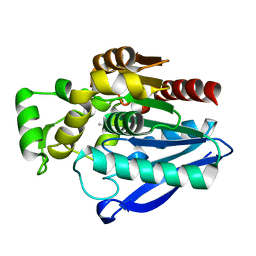 | | Crystal Structure of mtbMGL K74A (Closed Cap Conformation) | | Descriptor: | ISOPROPYL ALCOHOL, Monoacylglycerol lipase | | Authors: | Grininger, C, Aschauer, P, Pavkov-Keller, T, Oberer, M. | | Deposit date: | 2021-06-28 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Changes in the Cap of Rv0183/mtbMGL Modulate the Shape of the Binding Pocket.
Biomolecules, 11, 2021
|
|
7P0Y
 
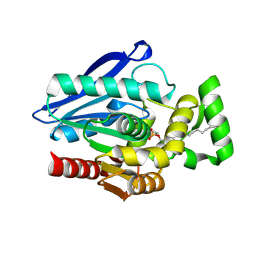 | | Crystal Structure of mtbMGL K74A (Substrate Analog Complex) | | Descriptor: | 1-[butyl(fluoranyl)phosphoryl]oxyhexadecane, Monoacylglycerol lipase | | Authors: | Grininger, C, Aschauer, P, Pavkov-Keller, T, Oberer, M. | | Deposit date: | 2021-06-30 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Changes in the Cap of Rv0183/mtbMGL Modulate the Shape of the Binding Pocket.
Biomolecules, 11, 2021
|
|
2H3A
 
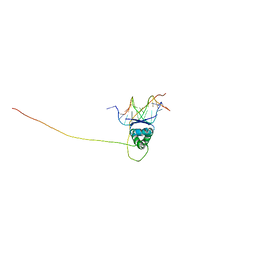 | | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA | | Descriptor: | 5'-D(P*AP*TP*AP*TP*GP*TP*AP*TP*AP*CP*CP*CP*G)-3', 5'-D(P*TP*CP*GP*GP*GP*TP*AP*TP*AP*CP*AP*TP*A)-3', CcdA | | Authors: | Madl, T, Van Melderen, L, Respondek, M, Oberer, M, Keller, W, Zangger, K. | | Deposit date: | 2006-05-22 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Nucleic Acid and Toxin Recognition of the Bacterial Antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2H3C
 
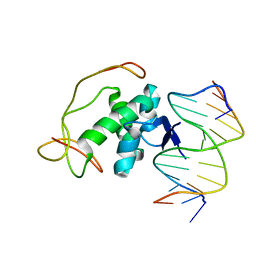 | | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA | | Descriptor: | 5'-D(P*AP*TP*AP*TP*GP*TP*AP*TP*AP*CP*CP*CP*G)-3', 5'-D(P*TP*CP*GP*GP*GP*TP*AP*TP*AP*CP*AP*TP*A)-3', CcdA | | Authors: | Madl, T, Van Melderen, L, Respondek, M, Oberer, M, Keller, W, Zangger, K. | | Deposit date: | 2006-05-22 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Nucleic Acid and Toxin Recognition of the Bacterial Antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
3RLI
 
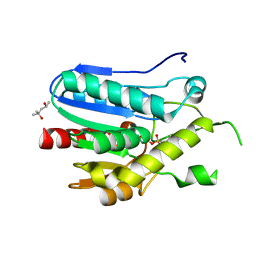 | | Crystal structure of monoacylglycerol lipase from Bacillus sp. H257 in complex with PMSF | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Thermostable monoacylglycerol lipase, phenylmethanesulfonic acid | | Authors: | Rengachari, S, Bezerra, G.A, Gruber, K, Oberer, M. | | Deposit date: | 2011-04-19 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | The structure of monoacylglycerol lipase from Bacillus sp. H257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes.
Biochim.Biophys.Acta, 1821, 2012
|
|
3RM3
 
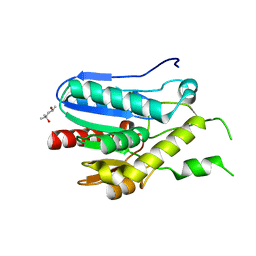 | | Crystal structure of monoacylglycerol lipase from Bacillus sp. H257 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Thermostable monoacylglycerol lipase | | Authors: | Rengachari, S, Bezerra, G.A, Gruber, K, Oberer, M. | | Deposit date: | 2011-04-20 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structure of monoacylglycerol lipase from Bacillus sp. H257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes.
Biochim.Biophys.Acta, 1821, 2012
|
|
4ZWN
 
 | | Crystal Structure of a Soluble Variant of the Monoglyceride Lipase from Saccharomyces Cerevisiae | | Descriptor: | Monoglyceride lipase, NITRATE ION, SODIUM ION, ... | | Authors: | Aschauer, P, Rengachari, S, Gruber, K, Oberer, M. | | Deposit date: | 2015-05-19 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae monoglyceride lipase Yju3p.
Biochim.Biophys.Acta, 1861, 2016
|
|
4ZXF
 
 | | Crystal Structure of a Soluble Variant of Monoglyceride Lipase from Saccharomyces Cerevisiae in Complex with a Substrate Analog | | Descriptor: | 1-{3-[(R)-hydroxy(octadecyloxy)phosphoryl]propyl}triaza-1,2-dien-2-ium, Monoglyceride lipase, NITRATE ION, ... | | Authors: | Aschauer, P, Lichtenegger, J, Rengachari, S, Gruber, K, Oberer, M. | | Deposit date: | 2015-05-20 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae monoglyceride lipase Yju3p.
Biochim.Biophys.Acta, 1861, 2016
|
|
5A4H
 
 | | Solution structure of the lipid droplet anchoring peptide of CGI-58 bound to DPC micelles | | Descriptor: | 1-ACYLGLYCEROL-3-PHOSPHATE O-ACYLTRANSFERASE ABHD5 | | Authors: | Boeszoermenyi, A, Arthanari, H, Wagner, G, Nagy, H.M, Zangger, K, Lindermuth, H, Oberer, M. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a Cgi-58 Motif Provides the Molecular Basis of Lipid Droplet Anchoring.
J.Biol.Chem., 290, 2015
|
|
4KE8
 
 | | Crystal structure of Monoglyceride lipase from Bacillus sp. H257 in complex with monopalmitoyl glycerol analogue | | Descriptor: | Thermostable monoacylglycerol lipase, tetradecyl hydrogen (R)-(3-azidopropyl)phosphonate | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
4KEA
 
 | | Crystal structure of D196N mutant of Monoglyceride lipase from Bacillus sp. H257 in space group P212121 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Thermostable monoacylglycerol lipase | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
4KE7
 
 | | Crystal structure of Monoglyceride lipase from Bacillus sp. H257 in complex with an 1-myristoyl glycerol analogue | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Thermostable monoacylglycerol lipase, dodecyl hydrogen (S)-(3-azidopropyl)phosphonate | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
4KE6
 
 | | Crystal structure D196N mutant of Monoglyceride lipase from Bacillus sp. H257 in complex with 1-rac-lauroyl glycerol | | Descriptor: | (2R)-2,3-dihydroxypropyl dodecanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, Thermostable monoacylglycerol lipase | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
4KE9
 
 | | Crystal structure of Monoglyceride lipase from Bacillus sp. H257 in complex with an 1-stearyol glycerol analogue | | Descriptor: | Thermostable monoacylglycerol lipase, hexadecyl hydrogen (R)-(3-azidopropyl)phosphonate | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
4MEL
 
 | | Crystal Structure of the human USP11 DUSP-UBL domains | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 11 | | Authors: | Harper, S, Gratton, H.E, Cornaciu, I, Oberer, M, Scott, D.J, Emsley, J, Dreveny, I. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Structure and Catalytic Regulatory Function of Ubiquitin Specific Protease 11 N-Terminal and Ubiquitin-like Domains.
Biochemistry, 53, 2014
|
|
4MEM
 
 | | Crystal Structure of the rat USP11 DUSP-UBL domains | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 11 | | Authors: | Harper, S, Gratton, H.E, Cornaciu, I, Oberer, M, Scott, D.J, Emsley, J, Dreveny, I. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure and Catalytic Regulatory Function of Ubiquitin Specific Protease 11 N-Terminal and Ubiquitin-like Domains.
Biochemistry, 53, 2014
|
|
1XC8
 
 | | CRYSTAL STRUCTURE COMPLEX BETWEEN THE WILD-TYPE LACTOCOCCUS LACTIS FPG (MUTM) AND A FAPY-DG CONTAINING DNA | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*(FOX)P*TP*TP*TP*CP*TP*CP*G)-3', 5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3', Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Coste, F, Ober, M, Carell, T, Boiteux, S, Zelwer, C, Castaing, B. | | Deposit date: | 2004-09-01 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the recognition of the FapydG lesion (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by formamidopyrimidine-DNA glycosylase.
J.Biol.Chem., 279, 2004
|
|
6FL1
 
 | | Crystal structure of the complex between the Lactococcus lactis FPG mutant T221P and a Fapy-dG containing DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*TP*TP(FOX)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Coste, F, Castaing, B, Ober, M, Carell, T. | | Deposit date: | 2018-01-25 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the complex between the Lactococcus lactis FPG mutant T221P and a Fapy-dG containing DNA
To Be Published
|
|
1U45
 
 | | 8oxoguanine at the pre-insertion site of the polymerase active site | | Descriptor: | DNA polymerase I, DNA primer strand, DNA template strand with 8-oxoguanine, ... | | Authors: | Hsu, G.W, Ober, M, Carell, T, Beese, L.S. | | Deposit date: | 2004-07-23 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase.
Nature, 431, 2004
|
|
