1SSN
 
 | | STAPHYLOKINASE, SAKSTAR VARIANT, NMR, 20 STRUCTURES | | Descriptor: | STAPHYLOKINASE | | Authors: | Ohlenschlager, O, Ramachandran, R, Guhrs, K.H, Schlott, B, Brown, L.R. | | Deposit date: | 1998-06-07 | | Release date: | 1998-12-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the plasminogen-activator protein staphylokinase.
Biochemistry, 37, 1998
|
|
1NU0
 
 | | Structure of the double mutant (L6M; F134M, SeMet form) of yqgF from Escherichia coli, a hypothetical protein | | Descriptor: | Hypothetical protein yqgF, SULFATE ION | | Authors: | Galkin, A, Sarikaya, E, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-30 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of yqgF from Escherichia coli, a hypothetical protein
To be Published
|
|
1A0D
 
 | | XYLOSE ISOMERASE FROM BACILLUS STEAROTHERMOPHILUS | | Descriptor: | MANGANESE (II) ION, XYLOSE ISOMERASE | | Authors: | Gallay, O, Chopra, R, Conti, E, Brick, P, Blow, D. | | Deposit date: | 1997-11-28 | | Release date: | 1998-06-03 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Class II Xylose Isomerases from Two Thermophiles and a Hyperthermophile
To be Published
|
|
1A4Q
 
 | | INFLUENZA VIRUS B/BEIJING/1/87 NEURAMINIDASE COMPLEXED WITH DIHYDROPYRAN-PHENETHYL-PROPYL-CARBOXAMIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-ACETYLAMINO-4-AMINO-6-(PHENETHYL-PROPYL-CARBAMOYL)-5,6-DIHYDRO-4H-PYRAN-2-CARBOXYLIC ACID, CALCIUM ION, ... | | Authors: | Cleasby, A, Singh, O, Skarzynski, T, Wonacott, A.J. | | Deposit date: | 1998-01-30 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dihydropyrancarboxamides related to zanamivir: a new series of inhibitors of influenza virus sialidases. 2. Crystallographic and molecular modeling study of complexes of 4-amino-4H-pyran-6-carboxamides and sialidase from influenza virus types A and B.
J.Med.Chem., 41, 1998
|
|
1NYP
 
 | | 4th LIM domain of PINCH protein | | Descriptor: | PINCH protein, ZINC ION | | Authors: | Velyvis, A, Vaynberg, J, Vinogradova, O, Zhang, Y, Wu, C, Qin, J. | | Deposit date: | 2003-02-13 | | Release date: | 2003-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and functional insights into PINCH LIM4 domain-mediated integrin signaling
Nat.Struct.Biol., 10, 2003
|
|
1NSK
 
 | |
1V33
 
 | | Crystal structure of DNA primase from Pyrococcus horikoshii | | Descriptor: | DNA primase small subunit, PHOSPHATE ION, ZINC ION | | Authors: | Ito, N, Nureki, O, Shirouzu, M, Yokoyama, S, Hanaoka, F, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-25 | | Release date: | 2004-03-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Pyrococcus horikoshii DNA primase-UTP complex: implications for the mechanism of primer synthesis.
Genes Cells, 8, 2003
|
|
1BCG
 
 | | SCORPION TOXIN BJXTR-IT | | Descriptor: | TOXIN BJXTR-IT | | Authors: | Oren, D, Froy, O, Amit, E, Kleinberger-Doron, N, Gurevitz, M, Shaanan, B. | | Deposit date: | 1998-04-29 | | Release date: | 1998-11-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An excitatory scorpion toxin with a distinctive feature: an additional alpha helix at the C terminus and its implications for interaction with insect sodium channels.
Structure, 6, 1998
|
|
1UOY
 
 | | The bubble protein from Penicillium brevicompactum Dierckx exudate. | | Descriptor: | BUBBLE PROTEIN | | Authors: | Olsen, J.G, Flensburg, C, Olsen, O, Seibold, M, Bricogne, G, Henriksen, A. | | Deposit date: | 2003-09-26 | | Release date: | 2003-11-04 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Solving the Structure of the Bubble Protein Using the Anomalous Sulfur Signal from Single-Crystal in-House Cu Kalpha Diffraction Data Only
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1BH8
 
 | | HTAFII18/HTAFII28 HETERODIMER CRYSTAL STRUCTURE | | Descriptor: | TAFII18, TAFII28 | | Authors: | Birck, C, Poch, O, Romier, C, Ruff, M, Mengus, G, Lavigne, A.-C, Davidson, I, Moras, D. | | Deposit date: | 1998-06-16 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family.
Cell(Cambridge,Mass.), 94, 1998
|
|
1QJV
 
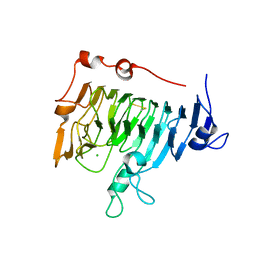 | | Pectin methylesterase PemA from Erwinia chrysanthemi | | Descriptor: | CHLORIDE ION, PECTIN METHYLESTERASE | | Authors: | Jenkins, J, Mayans, O, Smith, D, Worboys, K, Pickersgill, R. | | Deposit date: | 1999-07-05 | | Release date: | 2000-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Three-Dimensional Structure of Erwinia Chrysanthemi Pectin Methylesterase Reveals a Novel Esterase Active Site
J.Mol.Biol., 305, 2001
|
|
1UUU
 
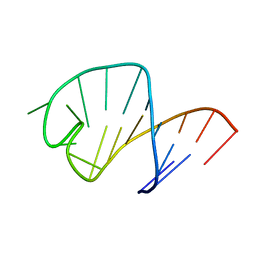 | | STRUCTURE OF AN RNA HAIRPIN LOOP WITH A 5'-CGUUUCG-3' LOOP MOTIF BY HETERONUCLEAR NMR SPECTROSCOPY AND DISTANCE GEOMETRY, 15 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*UP*AP*CP*GP*UP*UP*UP*CP*GP*UP*AP*CP*GP*CP*C)-3') | | Authors: | Sich, C, Ohlenschlager, O, Ramachandran, R, Gorlach, M, Brown, L.R. | | Deposit date: | 1997-08-12 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA hairpin loop with a 5'-CGUUUCG-3' loop motif by heteronuclear NMR spectroscopy and distance geometry.
Biochemistry, 36, 1997
|
|
8ERX
 
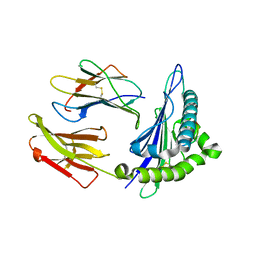 | | Structure of chimeric HLA-A*11:01-A*02:01 bound to HIV-1 RT peptide | | Descriptor: | Beta-2-microglobulin, HIV-1 RT, HLA-A*02:01 | | Authors: | Florio, T.J, Ani, O, Young, M.C, Mallik, L, Sgourakis, N.G. | | Deposit date: | 2022-10-13 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Decoupling peptide binding from T cell receptor recognition with engineered chimeric MHC-I molecules.
Front Immunol, 14, 2023
|
|
8ESH
 
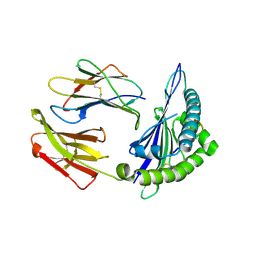 | | Structure of chimeric HLA-A*02:01 bound to CMV peptide | | Descriptor: | Beta-2-microglobulin, CMV peptide, HLA-A*02:01 | | Authors: | Florio, T.J, Ani, O, Young, M.C, Mallik, L, Sgourakis, N.G. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Decoupling peptide binding from T cell receptor recognition with engineered chimeric MHC-I molecules.
Front Immunol, 14, 2023
|
|
1V34
 
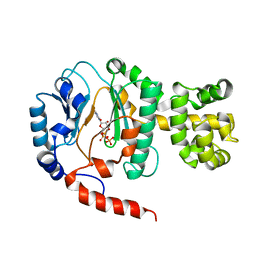 | | Crystal structure of Pyrococcus horikoshii DNA primase-UTP complex | | Descriptor: | DNA primase small subunit, URIDINE 5'-TRIPHOSPHATE, ZINC ION | | Authors: | Ito, N, Nureki, O, Shirouzu, M, Yokoyama, S, Hanaoka, F, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-25 | | Release date: | 2004-03-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Pyrococcus horikoshii DNA primase-UTP complex: implications for the mechanism of primer synthesis.
Genes Cells, 8, 2003
|
|
1R7A
 
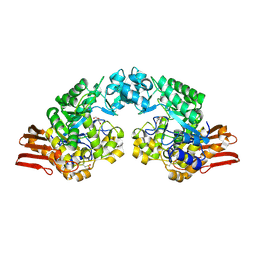 | | Sucrose Phosphorylase from Bifidobacterium adolescentis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, sucrose phosphorylase | | Authors: | Sprogoe, D, van den Broek, L.A.M, Mirza, O, Kastrup, J.S, Voragen, A.G.J, Gajhede, M, Skov, L.K. | | Deposit date: | 2003-10-21 | | Release date: | 2004-02-10 | | Last modified: | 2014-11-19 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of sucrose phosphorylase from Bifidobacterium adolescentis.
Biochemistry, 43, 2004
|
|
8EET
 
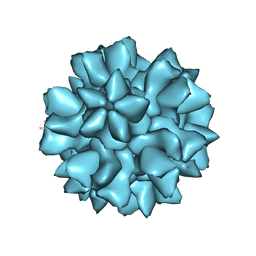 | |
8EEP
 
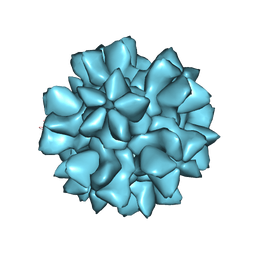 | |
8EUS
 
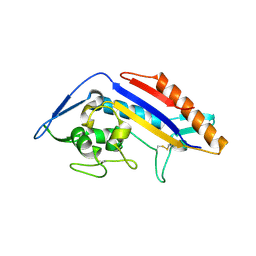 | | Crystal structure of NPC1 luminal domain C | | Descriptor: | NPC intracellular cholesterol transporter 1 | | Authors: | Odongo, L, Pornillos, O. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Purification and structure of luminal domain C of human Niemann-Pick C1 protein.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8EJL
 
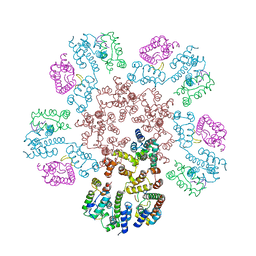 | | Structure of HIV-1 capsid declination in complex with CPSF6-FG peptide | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 6, HIV-1 capsid protein | | Authors: | Pornillos, O, Ganser-Pornillos, B.K, Schirra, R.T, dos Santos, N.F.B. | | Deposit date: | 2022-09-17 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A molecular switch modulates assembly and host factor binding of the HIV-1 capsid.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3TTA
 
 | |
1QGJ
 
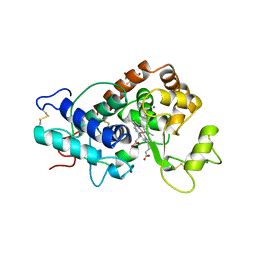 | | ARABIDOPSIS THALIANA PEROXIDASE N | | Descriptor: | CALCIUM ION, GLUTATHIONE, PEROXIDASE N, ... | | Authors: | Mirza, O, Oestergaard, L, Welinder, K.G, Henriksen, A, Gajhede, M. | | Deposit date: | 1999-04-29 | | Release date: | 2000-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Arabidopsis thaliana peroxidase N: structure of a novel neutral peroxidase.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1TFF
 
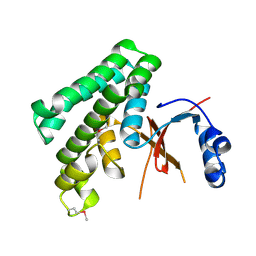 | | Structure of Otubain-2 | | Descriptor: | Ubiquitin thiolesterase protein OTUB2 | | Authors: | Nanao, M.H, Tcherniuk, S.O, Chroboczek, J, Dideberg, O, Dessen, A, Balakirev, M.Y. | | Deposit date: | 2004-05-27 | | Release date: | 2004-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human otubain 2.
Embo Rep., 5, 2004
|
|
1SU7
 
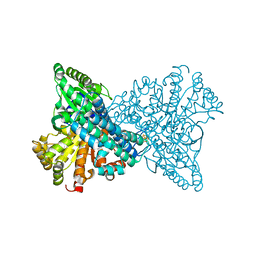 | | Carbon Monoxide Dehydrogenase from Carboxydothermus hydrogenoformans- DTT reduced state | | Descriptor: | Carbon monoxide dehydrogenase 2, FE(4)-NI(1)-S(5) CLUSTER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Dobbek, H, Svetlitchnyi, V, Liss, J, Meyer, O. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Carbon Monoxide Induced Decomposition of the Active Site [Ni-4Fe-5S] Cluster of CO Dehydrogenase
J.Am.Chem.Soc., 126, 2004
|
|
1T6C
 
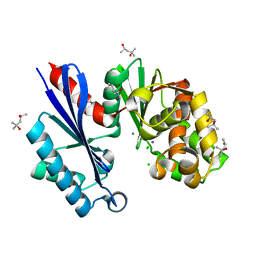 | | Structural characterization of the Ppx/GppA protein family: crystal structure of the Aquifex aeolicus family member | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kristensen, O, Laurberg, M, Liljas, A, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2004-05-06 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural characterization of the stringent response related exopolyphosphatase/guanosine pentaphosphate phosphohydrolase protein family
Biochemistry, 43, 2004
|
|
