1O91
 
 | | Crystal Structure of a Collagen VIII NC1 Domain Trimer | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, COLLAGEN ALPHA 1(VIII) CHAIN, SULFATE ION | | Authors: | Kvansakul, M, Bogin, O, Hohenester, E, Yayon, A. | | Deposit date: | 2002-12-10 | | Release date: | 2003-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Collagen Alpha1(Viii) Nc1 Trimer.
Matrix Biol., 22, 2003
|
|
1PR9
 
 | | Human L-Xylulose Reductase Holoenzyme | | Descriptor: | DIHYDROGENPHOSPHATE ION, L-XYLULOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | El-Kabbani, O, Ishikura, S, Darmanin, C, Carbone, V, Chung, R.P.-T, Usami, N, Hara, A. | | Deposit date: | 2003-06-20 | | Release date: | 2004-02-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of human L-xylulose reductase holoenzyme: probing the role of Asn107 with site-directed mutagenesis
Proteins, 55, 2004
|
|
3EYG
 
 | | Crystal structures of JAK1 and JAK2 inhibitor complexes | | Descriptor: | 3-{(3R,4R)-4-methyl-3-[methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]piperidin-1-yl}-3-oxopropanenitrile, Tyrosine-protein kinase | | Authors: | Williams, N.K, Bamert, R.S, Patell, O, Wang, C, Walden, P.M, Fantino, E. | | Deposit date: | 2008-10-20 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dissecting specificity in the Janus kinases: the structures of JAK-specific inhibitors complexed to the JAK1 and JAK2 protein tyrosine kinase domains.
J.Mol.Biol., 387, 2009
|
|
1IKC
 
 | | NMR Structure of alpha-Bungarotoxin | | Descriptor: | long neurotoxin 1 | | Authors: | Niccolai, N, Spiga, O, Ciutti, A. | | Deposit date: | 2001-05-03 | | Release date: | 2001-05-16 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of alpha-bungarotoxin free and bound to a mimotope of the nicotinic acetylcholine receptor.
Biochemistry, 41, 2002
|
|
1LCW
 
 | | streptavidin-homobiotin complex | | Descriptor: | HOMOBIOTIN, Streptavidin | | Authors: | Livnah, O, Pazy, Y, Bayer, E.A, Wilchek, M. | | Deposit date: | 2002-04-07 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand exchange between proteins: exchange of biotin and biotin derivatives between avidin and streptavidin
J.Biol.Chem., 277, 2002
|
|
1P0M
 
 | | Crystal structure of human butyryl cholinesterase in complex with a choline molecule | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nicolet, Y, Lockridge, O, Masson, P, Fontecilla-Camps, J.C, Nachon, F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of human butyrylcholinesterase and of its complexes with substrate and products.
J.Biol.Chem., 278, 2003
|
|
3UAG
 
 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Bertrand, J.A, Auger, G, Martin, L, Fanchon, E, Blanot, D, Le Beller, D, Van Heijenoort, J, Dideberg, O. | | Deposit date: | 1999-02-24 | | Release date: | 2000-02-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Determination of the MurD mechanism through crystallographic analysis of enzyme complexes.
J.Mol.Biol., 289, 1999
|
|
1PYZ
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF MIMOCHROME IV | | Descriptor: | CHLORIDE ION, CO(III)-(DEUTEROPORPHYRIN IX), MIMOCHROME IV, ... | | Authors: | Di Costanzo, L, Geremia, S, Randaccio, L, Nastri, F, Maglio, O, Lombardi, A, Pavone, V. | | Deposit date: | 2003-07-09 | | Release date: | 2004-12-14 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Miniaturized heme proteins: crystal structure of Co(III)-mimochrome IV.
J.Biol.Inorg.Chem., 9, 2004
|
|
1LEL
 
 | | The avidin BCAP complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin, E-AMINO BIOTINYL CAPROIC ACID | | Authors: | Pazy, Y, Kulik, T, Bayer, E.A, Wilchek, M, Livnah, O. | | Deposit date: | 2002-04-10 | | Release date: | 2002-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand exchange between proteins: exchange of biotin and biotin derivatives between avidin and streptavidin
J.Biol.Chem., 277, 2002
|
|
3IZD
 
 | | Model of the large subunit RNA expansion segment ES27L-out based on a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome. 3IZD is a small part (an expansion segment) which is in an alternative conformation to what is in already 3IZF. | | Descriptor: | rRNA expansion segment ES27L in an "out" conformation | | Authors: | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2010-10-13 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1LBU
 
 | | HYDROLASE METALLO (ZN) DD-PEPTIDASE | | Descriptor: | MURAMOYL-PENTAPEPTIDE CARBOXYPEPTIDASE, ZINC ION | | Authors: | Charlier, P, Wery, J.-P, Dideberg, O, Frere, J.-M. | | Deposit date: | 1996-03-16 | | Release date: | 1996-11-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Streptomyces Albus G D-Ala-A-Ala Carboxypeptidase
Handbook of Metalloproteins, 3, 2004
|
|
3S2R
 
 | | ATChloroNEET (H87C mutant) | | Descriptor: | AT5g51720/MIO24_14, FE2/S2 (INORGANIC) CLUSTER | | Authors: | livnah, O, Eisenberg-Domovich, Y, nechushtai, R. | | Deposit date: | 2011-05-17 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Arabidopsis thaliana ChloroNEET, a Member of the New NEET Family of Human Proteins, is Involved in Development, Senescence and Iron Metabolism.
To be Published
|
|
3S2Q
 
 | | The crystal structure of AT5g51720 (AT-NEET) | | Descriptor: | AT5g51720/MIO24_14, FE2/S2 (INORGANIC) CLUSTER, ZINC ION | | Authors: | Livnah, O, Eisenberg-Domovich, Y, Nechushtai, R. | | Deposit date: | 2011-05-17 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Arabidopsis thaliana ChloroNEET, a Member of the New NEET Family of Human Proteins, is Involved in Development, Senescence and Iron Metabolism.
To be Published
|
|
1PMD
 
 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) | | Descriptor: | PEPTIDOGLYCAN SYNTHESIS MULTIFUNCTIONAL ENZYME | | Authors: | Pares, S, Mouz, N, Dideberg, O. | | Deposit date: | 1996-02-05 | | Release date: | 1997-02-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme.
Nat.Struct.Biol., 3, 1996
|
|
8EUS
 
 | | Crystal structure of NPC1 luminal domain C | | Descriptor: | NPC intracellular cholesterol transporter 1 | | Authors: | Odongo, L, Pornillos, O. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Purification and structure of luminal domain C of human Niemann-Pick C1 protein.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8EJL
 
 | | Structure of HIV-1 capsid declination in complex with CPSF6-FG peptide | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 6, HIV-1 capsid protein | | Authors: | Pornillos, O, Ganser-Pornillos, B.K, Schirra, R.T, dos Santos, N.F.B. | | Deposit date: | 2022-09-17 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A molecular switch modulates assembly and host factor binding of the HIV-1 capsid.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2BMI
 
 | | METALLO-BETA-LACTAMASE | | Descriptor: | PROTEIN (CLASS B BETA-LACTAMASE), SODIUM ION, ZINC ION | | Authors: | Carfi, A, Duee, E, Dideberg, O. | | Deposit date: | 1998-09-17 | | Release date: | 1998-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of the ZnII beta-lactamase from Bacteroides fragilis in an orthorhombic crystal form.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
6GQJ
 
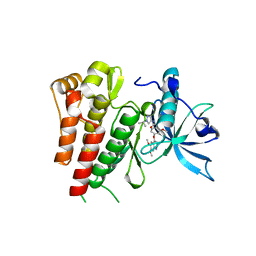 | | Crystal structure of human c-KIT kinase domain in complex with AZD3229-analogue (compound 18) | | Descriptor: | 2-[4-(6,7-dimethoxyquinazolin-4-yl)oxy-2-methoxy-phenyl]-~{N}-(1-propan-2-ylpyrazol-4-yl)ethanamide, Mast/stem cell growth factor receptor Kit | | Authors: | Schimpl, M, Hardy, C.J, Ogg, D.J, Overman, R.C, Packer, M.J, Kettle, J.G, Anjum, R, Barry, E, Bhavsar, D, Brown, C, Campbell, A, Goldberg, K, Grondine, M, Guichard, S, Hunt, T, Jones, O, Li, X, Moleva, O, Pearson, S, Shao, W, Smith, A, Smith, J, Stead, D, Stokes, S, Tucker, M, Ye, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery of N-(4-{[5-Fluoro-7-(2-methoxyethoxy)quinazolin-4-yl]amino}phenyl)-2-[4-(propan-2-yl)-1 H-1,2,3-triazol-1-yl]acetamide (AZD3229), a Potent Pan-KIT Mutant Inhibitor for the Treatment of Gastrointestinal Stromal Tumors.
J. Med. Chem., 61, 2018
|
|
8G1Y
 
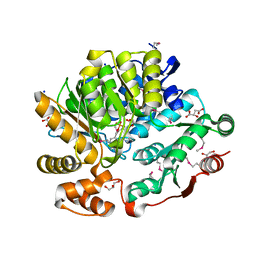 | | Crystal Structure of the Threonine Synthase from Streptococcus pneumoniae in complex with Pyridoxal 5-phosphate. | | Descriptor: | CHLORIDE ION, D-MALATE, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-02-03 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal Structure of the Threonine Synthase from Streptococcus pneumoniae in complex with Pyridoxal 5-phosphate.
To Be Published
|
|
8G22
 
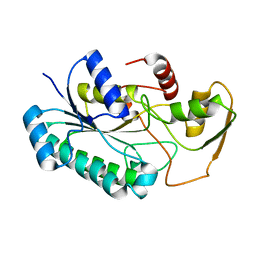 | | Crystal Structure of the dTDP-4-dehydrorhamnose Reductase from Streptococcus pneumoniae. | | Descriptor: | dTDP-4-dehydrorhamnose reductase | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-02-03 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal Structure of the dTDP-4-dehydrorhamnose Reductase from Streptococcus pneumoniae.
To Be Published
|
|
8G28
 
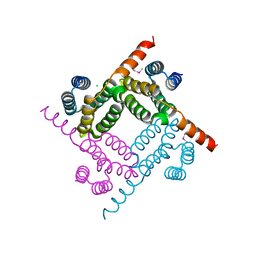 | | Crystal Structure of the C-terminal Fragment of AAA ATPase from Streptococcus pneumoniae. | | Descriptor: | ATPase, AAA family, CHLORIDE ION | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-02-03 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of the C-terminal Fragment of AAA ATPase from Streptococcus pneumoniae.
To Be Published
|
|
6GQO
 
 | | Crystal structure of human KDR (VEGFR2) kinase domain in complex with AZD3229-analogue (compound 18) | | Descriptor: | 2-[4-(6,7-dimethoxyquinazolin-4-yl)oxy-2-methoxy-phenyl]-~{N}-(1-propan-2-ylpyrazol-4-yl)ethanamide, Vascular endothelial growth factor receptor 2 | | Authors: | Ogg, D.J, Schimpl, M, Hardy, C.J, Overman, R.C, Packer, M.J, Kettle, J.G, Anjum, R, Barry, E, Bhavsar, D, Brown, C, Campbell, A, Goldberg, K, Grondine, M, Guichard, S, Hunt, T, Jones, O, Li, X, Moleva, O, Pearson, S, Shao, W, Smith, A, Smith, J, Stead, D, Stokes, S, Tucker, M, Ye, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of N-(4-{[5-Fluoro-7-(2-methoxyethoxy)quinazolin-4-yl]amino}phenyl)-2-[4-(propan-2-yl)-1 H-1,2,3-triazol-1-yl]acetamide (AZD3229), a Potent Pan-KIT Mutant Inhibitor for the Treatment of Gastrointestinal Stromal Tumors.
J. Med. Chem., 61, 2018
|
|
2WPS
 
 | | Salmonella enterica SadA 483-523 fused to GCN4 adaptors (SadAK3b-V2, out-of-register fusion) | | Descriptor: | CHLORIDE ION, TRIMERIC AUTOTRANSPORTER ADHESIN FRAGMENT | | Authors: | Hartmann, M.D, Ridderbusch, O, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2009-08-09 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Coiled-Coil Motif that Sequesters Ions to the Hydrophobic Core.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8FMF
 
 | | Structure of CBASS Cap5 from Pseudomonas syringae as an activated tetramer with the cyclic dinucleotide 3'2'-c-diAMP ligand (1 tetramer in the AU) | | Descriptor: | Cyclic (adenosine-(2'-5')-monophosphate-adenosine-(3'-5')-monophosphate, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Rechkoblit, O, Kreitler, D.F, Aggarwal, A.K. | | Deposit date: | 2022-12-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activation of CBASS Cap5 endonuclease immune effector by cyclic nucleotides.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FM1
 
 | |
