1KVD
 
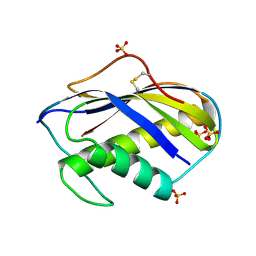 | | KILLER TOXIN FROM HALOTOLERANT YEAST | | Descriptor: | SMK TOXIN, SULFATE ION | | Authors: | Kashiwagi, T, Kunishima, N, Suzuki, C, Tsuchiya, F, Nikkuni, S, Arata, Y, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-04-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The novel acidophilic structure of the killer toxin from halotolerant yeast demonstrates remarkable folding similarity with a fungal killer toxin.
Structure, 5, 1997
|
|
1X01
 
 | |
1KVE
 
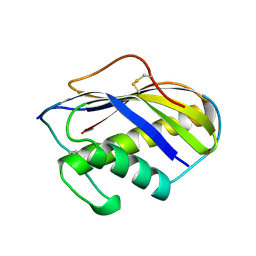 | | KILLER TOXIN FROM HALOTOLERANT YEAST | | Descriptor: | SMK TOXIN | | Authors: | Kashiwagi, T, Kunishima, N, Suzuki, C, Tsuchiya, F, Nikkuni, S, Arata, Y, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-04-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The novel acidophilic structure of the killer toxin from halotolerant yeast demonstrates remarkable folding similarity with a fungal killer toxin.
Structure, 5, 1997
|
|
1V8G
 
 | |
1VBI
 
 | |
1V7Q
 
 | |
1WWZ
 
 | |
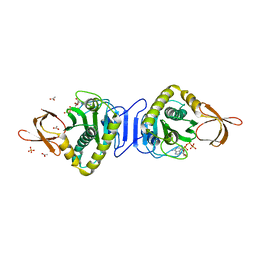
1V37
 
 | | Crystal structure of phosphoglycerate mutase from Thermus thermophilus HB8 | | Descriptor: | GLYCEROL, phosphoglycerate mutase | | Authors: | Sugahara, M, Yokoyama, S, Kuramitsu, S, Miyano, M, Iizuka, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-29 | | Release date: | 2004-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of phosphoglycerate mutase from Thermus thermophilus HB8
To be Published
|
|
1V8C
 
 | |
1V70
 
 | |
1V6S
 
 | |
1YYA
 
 | |
1ZIA
 
 | | OXIDIZED PSEUDOAZURIN | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Inoue, T, Nishio, N, Hamanaka, S, Shimomura, T, Harada, S, Suzuki, S, Kohzuma, T, Shidara, S, Iwasaki, H, Kai, Y. | | Deposit date: | 1996-04-09 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystallization and preliminary X-ray studies on pseudoazurin from Achromobacter cycloclastes IAM1013.
J.Biochem.(Tokyo), 114, 1993
|
|
1ZIB
 
 | | REDUCED PSEUDOAZURIN | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Inoue, T, Nishio, N, Hamanaka, S, Shimomura, T, Harada, S, Suzuki, S, Kohzuma, T, Shidara, S, Iwasaki, H, Kai, Y. | | Deposit date: | 1996-04-09 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization and preliminary X-ray studies on pseudoazurin from Achromobacter cycloclastes IAM1013.
J.Biochem.(Tokyo), 114, 1993
|
|
1ZZG
 
 | |
7CJU
 
 | |
1ZJJ
 
 | |
2CQZ
 
 | |
2CSU
 
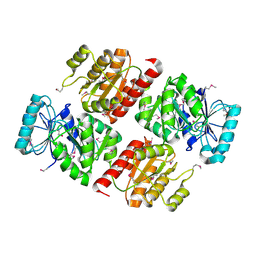 | |
6KIQ
 
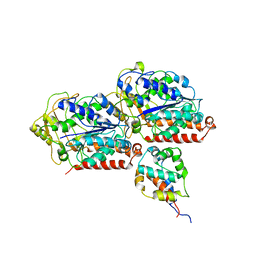 | | Complex of yeast cytoplasmic dynein MTBD-High and MT with DTT | | Descriptor: | Alpha tubulin, Dynein heavy chain, cytoplasmic, ... | | Authors: | Komori, Y, Nishida, N, Shimada, I, Kikkawa, M. | | Deposit date: | 2019-07-19 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis for two-way communication between dynein and microtubules.
Nat Commun, 11, 2020
|
|
6KIO
 
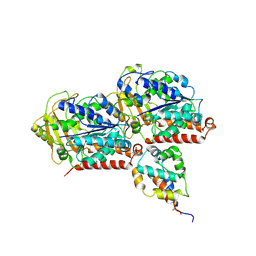 | | Complex of yeast cytoplasmic dynein MTBD-High and MT without DTT | | Descriptor: | Dynein heavy chain, cytoplasmic, Tubulin alpha-1A chain, ... | | Authors: | Komori, Y, Nishida, N, Shimada, I, Kikkawa, M. | | Deposit date: | 2019-07-19 | | Release date: | 2020-03-04 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Structural basis for two-way communication between dynein and microtubules.
Nat Commun, 11, 2020
|
|
2Z4F
 
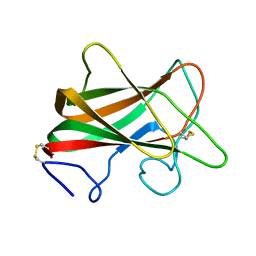 | | Solution structure of the Discoidin Domain of DDR2 | | Descriptor: | Discoidin domain-containing receptor 2 | | Authors: | Ichikawa, O, Osawa, M, Nishida, N, Goshima, N, Nomura, N, Shimada, I. | | Deposit date: | 2007-06-16 | | Release date: | 2007-09-04 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the collagen-binding mode of discoidin domain receptor 2
Embo J., 26, 2007
|
|
2Z6R
 
 | | Crystal structure of Lys49 to Arg mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Krishna Swamy, B.S, Simanshu, D.K, Murthy, M.R.N, Kunishima, N. | | Deposit date: | 2007-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
2DM9
 
 | |
2DMA
 
 | |
