6VZC
 
 | |
2LV0
 
 | |
17RA
 
 | |
2LUN
 
 | |
5UF3
 
 | |
4PDB
 
 | |
2AWQ
 
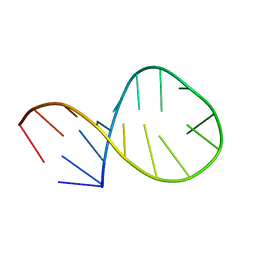 | |
2LA9
 
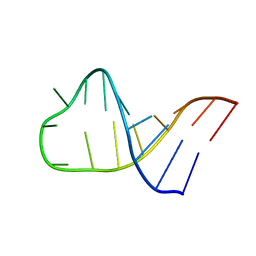 | | NMR structure of Pseudouridine_ASL_Tyr | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*AP*CP*UP*GP*UP*AP*AP*AP*(PSU)P*CP*CP*CP*C)-3') | | Authors: | Denmon, A.P, Wang, J, Nikonowicz, E.P. | | Deposit date: | 2011-03-09 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformation Effects of Base Modification on the Anticodon Stem-Loop of Bacillus subtilis tRNA(Tyr).
J.Mol.Biol., 412, 2011
|
|
2LBL
 
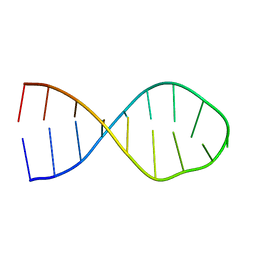 | |
2LAC
 
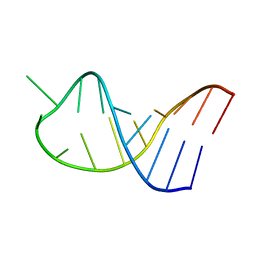 | | NMR structure of unmodified_ASL_Tyr | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*AP*CP*UP*GP*UP*AP*AP*AP*UP*CP*CP*CP*C)-3') | | Authors: | Denmon, A.P, Wang, J, Nikonowicz, E.P. | | Deposit date: | 2011-03-10 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformation Effects of Base Modification on the Anticodon Stem-Loop of Bacillus subtilis tRNA(Tyr).
J.Mol.Biol., 412, 2011
|
|
2LBJ
 
 | |
1KKA
 
 | |
1JWC
 
 | |
1J4Y
 
 | |
1JU7
 
 | |
2LBK
 
 | |
2LBR
 
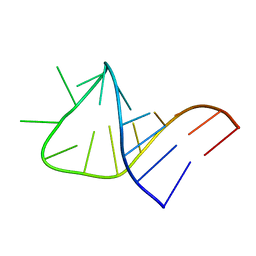 | |
2LBQ
 
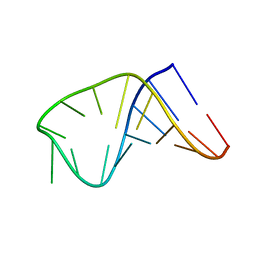 | | NMR structure of i6A37_tyrASL | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*AP*CP*UP*GP*UP*AP*(6IA)P*AP*UP*CP*CP*CP*C)-3') | | Authors: | Denmon, A.P, Wang, J, Nikonowicz, E.P. | | Deposit date: | 2011-04-05 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformation Effects of Base Modification on the Anticodon Stem-Loop of Bacillus subtilis tRNA(Tyr).
J.Mol.Biol., 412, 2011
|
|
1D0T
 
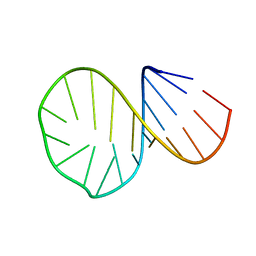 | |
1D0U
 
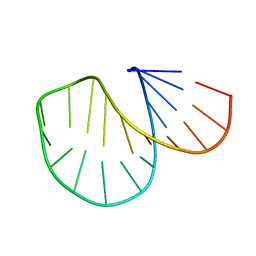 | |
1BGZ
 
 | |
