6TTB
 
 | | Crystal structure of NAD-dependent formate dehydrogenase from Staphylococcus aureus in complex with NAD | | Descriptor: | Formate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Boyko, K.M, Pometun, A.A, Nikolaeva, A.Y, Kargov, I.S, Yurchenko, T.S, Savin, S.S, Popov, V.O, Tishkov, V.I. | | Deposit date: | 2019-12-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of NAD-dependent formate dehydrogenase from Staphylococcus aureus in complex with NAD
To Be Published
|
|
7Q6B
 
 | | mRubyFT/S148I, a mutant of blue-to-red fluorescent timer in its blue state | | Descriptor: | mRubyFT S148I, a mutant of blue-to-red fluorescent timer | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Dorovatovskii, P.V, Khrenova, M.G, Subach, O.M, Popov, V.O, Subach, F.M. | | Deposit date: | 2021-11-06 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Combined Structural and Computational Study of the mRubyFT Fluorescent Timer Locked in Its Blue Form.
Int J Mol Sci, 24, 2023
|
|
8C0N
 
 | | Crystal structure of the red form of the mTagFT fluorescent timer | | Descriptor: | Blue-to-red TagFT fluorescent timer | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Agapova, Y.K, Subach, O.M, Popov, V.O, Subach, F.V. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Blue-to-Red TagFT, mTagFT, mTsFT, and Green-to-FarRed mNeptusFT2 Proteins, Genetically Encoded True and Tandem Fluorescent Timers.
Int J Mol Sci, 24, 2023
|
|
6THQ
 
 | | Crystal structure of branched-chain aminotransferase from thermophilic archaea Thermoproteus uzoniensis with norvaline | | Descriptor: | 2-[O-PHOSPHONOPYRIDOXYL]-AMINO-PENTANOIC ACID, Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2019-11-21 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Three-Dimensional Structure of Branched-Chain Amino Acid Transaminase from Thermoproteus uzoniensis in Complex with L-Norvaline
Crystallography Reports, 2020
|
|
8YTR
 
 | | The structure of Cu(II)-CopC from Thioalkalivibrio paradoxus | | Descriptor: | COPPER (II) ION, CopC domain-containing protein, DI(HYDROXYETHYL)ETHER | | Authors: | Kulikova, O.G, Solovieva, A.Y, Varfolomeeva, L.A, Dergousova, N.I, Nikolaeva, A.Y, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Cu(II)-CopC from Thioalkalivibrio paradoxus
To Be Published
|
|
5IJG
 
 | | Crystal structure of O-acetylhomoserine sulfhydrolase from Brucella melitensis at 2.0 A resolution | | Descriptor: | Cys/Met metabolism pyridoxal-phosphate-dependent enzyme, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Koolikova, V.V, Kotlov, M.I, Demidkina, T.V, Popov, V.O. | | Deposit date: | 2016-03-02 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of O-acetylhomoserine sulfhydrolase from Brucella melitensis at 2.0 A resolution
To Be Published
|
|
5JFQ
 
 | | Geranylgeranyl Pyrophosphate Synthetase from archaeon Geoglobus acetivorans | | Descriptor: | Geranylgeranyl Pyrophosphate Synthetase | | Authors: | Petrova, T, Boyko, K.M, Nikolaeva, A.Y, Stekhanova, T.N, Mardanov, A.V, Rakitin, A.L, Ravin, N.V, Popov, V.O. | | Deposit date: | 2016-04-19 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural characterization of geranylgeranyl pyrophosphate synthase GACE1337 from the hyperthermophilic archaeon Geoglobus acetivorans.
Extremophiles, 22, 2018
|
|
9J0V
 
 | | Crystal structure of monomeric PLP-dependent transaminase from Desulfobacula toluolica in P 21 21 21 space group | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dat: predicted D-alanine aminotransferase, GLYCEROL, ... | | Authors: | Matyuta, I.O, Bakunova, A.K, Nikolaeva, A.Y, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-08-03 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | High/low resolution monomeric PLP-dependent transaminase from Desulfobacula toluolica
To Be Published
|
|
9J0U
 
 | | Crystal structure of monomeric PLP-dependent transaminase from Desulfobacula toluolica in F 41 3 2 space group | | Descriptor: | Dat: predicted D-alanine aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Bakunova, A.K, Nikolaeva, A.Y, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-08-03 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | High/low resolution monomeric PLP-dependent transaminase from Desulfobacula toluolica
To Be Published
|
|
6ER1
 
 | | Crystal structure of BTB-domain of CP190 from D.melanogaster at high resolution | | Descriptor: | Centrosome-associated zinc finger protein CP190, PHOSPHATE ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Bonchuk, A.N, Kachalova, G.S, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2017-10-16 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Purification, Isolation, Crystallization, and Preliminary X-ray Diffraction Study of the BTB Domain of the Centrosomal Protein 190 from Drosophila Melanogaster
Crystallography Reports, 62, 2017
|
|
6ERK
 
 | | Crystal structure of diaminopelargonic acid aminotransferase from Psychrobacter cryohalolentis | | Descriptor: | 1,2-ETHANEDIOL, Aminotransferase, GLYCEROL, ... | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Bezsudnova, E.Y, Stekhanova, T.N, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2017-10-18 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Diaminopelargonic acid transaminase from Psychrobacter cryohalolentis is active towards (S)-(-)-1-phenylethylamine, aldehydes and alpha-diketones.
Appl. Microbiol. Biotechnol., 102, 2018
|
|
7B1X
 
 | | Crystal structure of cold-active esterase PMGL3 from permafrost metagenomic library | | Descriptor: | esterase PMGL3 | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Petrovskaya, L.E, Kryukova, M.V, Kryukova, E.A, Korzhenevsky, D.A, Lomakina, G.Y, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Popov, V.O. | | Deposit date: | 2020-11-25 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Characterization of a Cold-Active PMGL3 Esterase with Unusual Oligomeric Structure.
Biomolecules, 11, 2021
|
|
7ARZ
 
 | | Ternary complex of NAD-dependent formate dehydrogenase from Physcomitrium patens | | Descriptor: | AZIDE ION, Formate dehydrogenase, mitochondrial, ... | | Authors: | Goryaynova, D.A, Nikolaeva, A.Y, Pometun, A.A, Savin, S.S, Parshin, P.D, Popov, V.O, Tishkov, V.I, Boyko, K.M. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ternary complex of NAD-dependent formate dehydrogenase from Physcomitrium patens
To Be Published
|
|
5MWC
 
 | | Crystal structure of the genetically-encoded green calcium indicator NTnC in its calcium bound state | | Descriptor: | CALCIUM ION, genetically-encoded green calcium indicator NTnC | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Rakitina, T.V, Popov, V.O, Subach, O.M, Barykina, N.V, Subach, F.V. | | Deposit date: | 2017-01-18 | | Release date: | 2018-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Enchanced variant of genetically-encoded green calcium indicator NTnC
To Be Published
|
|
6Q8E
 
 | | Crystal structure of branched-chain amino acid aminotransferase from Thermobaculum terrenum in PMP-form | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Branched-chain-amino-acid aminotransferase, CHLORIDE ION | | Authors: | Boyko, K.M, Bezsudnova, E.Y, Nikolaeva, A.Y, Zeifman, Y.S, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2018-12-14 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical and structural insights into PLP fold type IV transaminase from Thermobaculum terrenum.
Biochimie, 158, 2018
|
|
6QIN
 
 | | CRYSTAL STRUCTURE OF THE PMGL2 ESTERASE FROM PERMAFROST METAGENOMIC LIBRARY | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PMGL2 | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Kryukova, M.V, Petrovskaya, L.E, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Popov, V.O. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of PMGL2 esterase from the hormone-sensitive lipase family with GCSAG motif around the catalytic serine.
Plos One, 15, 2020
|
|
6QLA
 
 | | CRYSTAL STRUCTURE OF THE PMGL2 ESTERASE (point mutant 1) FROM PERMAFROST METAGENOMIC LIBRARY | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Boyko, K.M, Garsia, D, Nikolaeva, A.Y, Korzhenevskiy, D.A, Kryukova, M.V, Petrovskaya, L.E, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Popov, V.O. | | Deposit date: | 2019-01-31 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of PMGL2 esterase from the hormone-sensitive lipase family with GCSAG motif around the catalytic serine.
Plos One, 15, 2020
|
|
8YQ4
 
 | | Structure of mBaoJin2 | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Minyaev, M.E, Kuzmicheva, T.P, Vlaskina, A.V, Popov, V.O, Pyatkevich, K.D, Subach, F.V. | | Deposit date: | 2024-03-19 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of mBaoJin2
To Be Published
|
|
8YRV
 
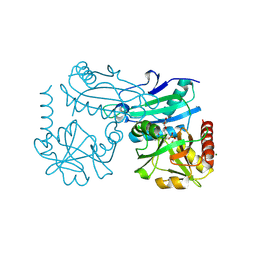 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis complexed with 3-aminooxypropionic acid | | Descriptor: | 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV, MAGNESIUM ION | | Authors: | Matyuta, I.O, Bakunova, A.K, Nikolaeva, A.Y, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-03-21 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis complexed with 3-aminooxypropionic acid
To Be Published
|
|
6FP5
 
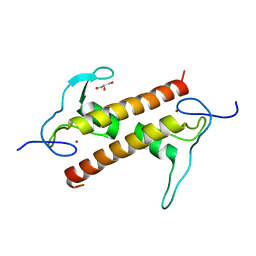 | | Crystal structure of ZAD-domain of CG2712 protein from D.melanogaster | | Descriptor: | CG2712, GLYCEROL, ZINC ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Bonchuk, A.N, Kachalova, G.S, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2018-02-09 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of diversity and homodimerization specificity of zinc-finger-associated domains in Drosophila.
Nucleic Acids Res., 49, 2021
|
|
6ET6
 
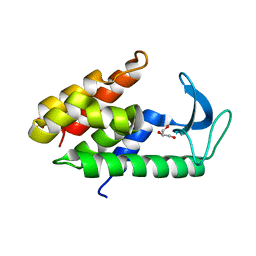 | | Crystal structure of muramidase from Acinetobacter baumannii AB 5075UW prophage | | Descriptor: | GLYCEROL, Lysozyme, SULFATE ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Sykilinda, N.N, Shneider, M.M, Miroshnikov, K.A, Popov, V.O. | | Deposit date: | 2017-10-25 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of anAcinetobacterBroad-Range Prophage Endolysin Reveals a C-Terminal alpha-Helix with the Proposed Role in Activity against Live Bacterial Cells.
Viruses, 10, 2018
|
|
6FUC
 
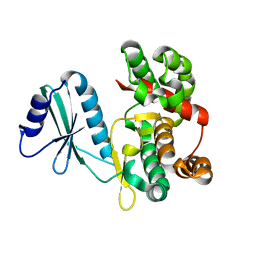 | | Structure of aminoglycoside phosphotransferase APH(3'')-Id from Streptomyces rimosus ATCC10970 | | Descriptor: | Aminoglycoside phosphotransferase | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Alekseeva, M.G, Mavletova, D.A, Zakharevich, N.V, Rudakova, N.N, Danilenko, V.N, Popov, V.O. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Identification, functional and structural characterization of novel aminoglycoside phosphotransferase APH(3′′)-Id from Streptomyces rimosus subsp. rimosus ATCC 10970.
Arch.Biochem.Biophys., 671, 2019
|
|
6FUX
 
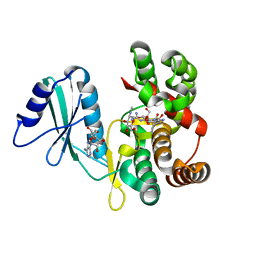 | | Structure of aminoglycoside phosphotransferase APH(3'')-Id from Streptomyces rimosus ATCC10970 in complex with ADP and streptomycin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aminoglycoside phosphotransferase, GLYCEROL, ... | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Alekseeva, M.G, Mavletova, D.A, Zakharevich, N.V, Rudakova, N.N, Danilenko, V.N, Popov, V.O. | | Deposit date: | 2018-02-28 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification, functional and structural characterization of novel aminoglycoside phosphotransferase APH(3′′)-Id from Streptomyces rimosus subsp. rimosus ATCC 10970.
Arch.Biochem.Biophys., 671, 2019
|
|
6GKR
 
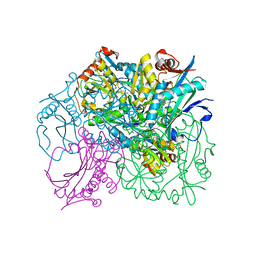 | | Crystal structure of branched-chain amino acid aminotransferase from Thermobaculum terrenum in PLP-form (holo-form) | | Descriptor: | ACETATE ION, Branched-chain-amino-acid aminotransferase, CHLORIDE ION, ... | | Authors: | Boyko, K.M, Bezsudnova, E.Y, Nikolaeva, A.Y, Zeifman, Y.S, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2018-05-21 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Biochemical and structural insights into PLP fold type IV transaminase from Thermobaculum terrenum.
Biochimie, 158, 2018
|
|
6H65
 
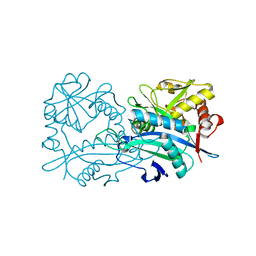 | | Crystal structure of the branched-chain-amino-acid aminotransferase from Haliangium ochraceum | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Boyko, K.M, Timofeev, V.I, Bezsudnova, E.Y, Nikolaeva, A.Y, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2018-07-26 | | Release date: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the branched-chain-amino-acid aminotransferase from Haliangium ochraceum
To Be Published
|
|
