8Q9S
 
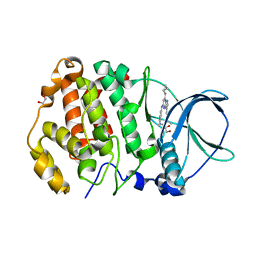 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 GENE PRODUCT) IN COMPLEX WITH THE INHIBITOR SGC-CK2-1 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha', ~{N}-[5-[[3-cyano-7-(cyclopropylamino)-3~{H}-pyrazolo[1,5-a]pyrimidin-5-yl]amino]-2-methyl-phenyl]propanamide | | Authors: | Werner, C, Lindenblatt, D, Niefind, K. | | Deposit date: | 2023-08-21 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.352 Å) | | Cite: | Discovery and Exploration of Protein Kinase CK2 Binding Sites Using CK2alpha Cys336Ser as an Exquisite Crystallographic Tool
Kinases Phosphatases, 2023
|
|
2NLO
 
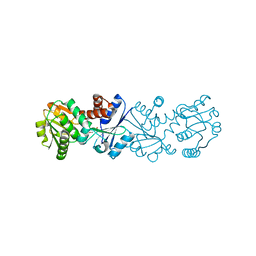 | |
5ONI
 
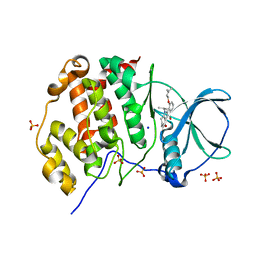 | | LOW-SALT STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR 4P | | Descriptor: | 1,4-BUTANEDIOL, 4-(3-methylbut-2-enoxy)-5-propan-2-yl-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, CHLORIDE ION, ... | | Authors: | Hochscherf, J, Lindenblatt, D, Witulski, B, Birus, R, Aichele, D, Marminon, C, Bouaziz, Z, Le Borgne, M, Jose, J, Niefind, K. | | Deposit date: | 2017-08-03 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected Binding Mode of a Potent Indeno[1,2-b]indole-Type Inhibitor of Protein Kinase CK2 Revealed by Complex Structures with the Catalytic Subunit CK2 alpha and Its Paralog CK2 alpha '.
Pharmaceuticals (Basel), 10, 2017
|
|
5OOI
 
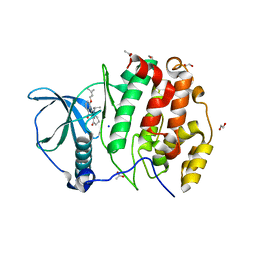 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA') IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR 4P | | Descriptor: | 1,2-ETHANEDIOL, 4-(3-methylbut-2-enoxy)-5-propan-2-yl-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, ACETATE ION, ... | | Authors: | Hochscherf, J, Lindenblatt, D, Witulski, B, Birus, R, Aichele, D, Marminon, C, Bouaziz, Z, Le Borgne, M, Jose, J, Niefind, K. | | Deposit date: | 2017-08-07 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Unexpected Binding Mode of a Potent Indeno[1,2-b]indole-Type Inhibitor of Protein Kinase CK2 Revealed by Complex Structures with the Catalytic Subunit CK2 alpha and Its Paralog CK2 alpha '.
Pharmaceuticals (Basel), 10, 2017
|
|
1DXY
 
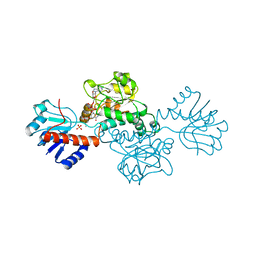 | | STRUCTURE OF D-2-HYDROXYISOCAPROATE DEHYDROGENASE | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, D-2-HYDROXYISOCAPROATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Dengler, U, Niefind, K, Kiess, M, Schomburg, D. | | Deposit date: | 1996-08-13 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of a ternary complex of D-2-hydroxyisocaproate dehydrogenase from Lactobacillus casei, NAD+ and 2-oxoisocaproate at 1.9 A resolution.
J.Mol.Biol., 267, 1997
|
|
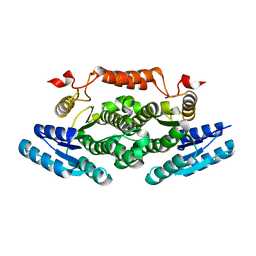
6ZE0
 
 | |
6ZDZ
 
 | |
5N9L
 
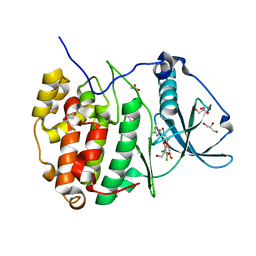 | | Crystal structure of human Protein kinase CK2 catalytic subunit in complex with the ATP-competitive dibenzofuran inhibitor TF (4b) | | Descriptor: | (4~{Z})-6,7-bis(chloranyl)-4-[[(4-methylphenyl)amino]methylidene]-8-oxidanyl-1,2-dihydrodibenzofuran-3-one, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Schnitzler, A, Gratz, A, Bollacke, A, Weyrich, M, Kucklaender, U, Wuensch, B, Goetz, C, Niefind, K, Jose, J. | | Deposit date: | 2017-02-25 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A pi-Halogen Bond of Dibenzofuranones with the Gatekeeper Phe113 in Human Protein Kinase CK2 Leads to Potent Tight Binding Inhibitors.
Pharmaceuticals, 11, 2018
|
|
5N9K
 
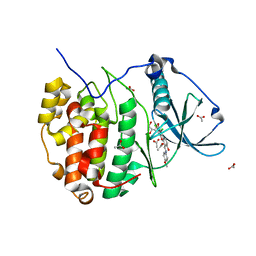 | | Crystal structure of human Protein kinase CK2 catalytic subunit in complex with the ATP-competitive, tight-binding dibenzofuran inhibitor TF107 (5) | | Descriptor: | 1,3-bis(chloranyl)-6-[(~{E})-(4-methoxyphenyl)iminomethyl]dibenzofuran-2,7-diol, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Schnitzler, A, Gratz, A, Bollacke, A, Weyrich, M, Kucklaender, U, Wuensch, B, Goetz, C, Niefind, K, Jose, J. | | Deposit date: | 2017-02-25 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.643 Å) | | Cite: | A pi-Halogen Bond of Dibenzofuranones with the Gatekeeper Phe113 in Human Protein Kinase CK2 Leads to Potent Tight Binding Inhibitors.
Pharmaceuticals, 11, 2018
|
|
5N9N
 
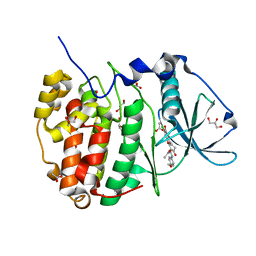 | | Crystal structure of human Protein kinase CK2 catalytic subunit in complex with the ATP-competitive, tight-binding dibenzofuran inhibitor TF85 (4a) | | Descriptor: | (4~{Z})-7,9-bis(chloranyl)-4-[[(4-methoxyphenyl)amino]methylidene]-8-oxidanyl-1,2-dihydrodibenzofuran-3-one, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Schnitzler, A, Gratz, A, Bollacke, A, Weyrich, M, Kucklaender, U, Wuensch, B, Goetz, C, Niefind, K, Jose, J. | | Deposit date: | 2017-02-25 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | A pi-Halogen Bond of Dibenzofuranones with the Gatekeeper Phe113 in Human Protein Kinase CK2 Leads to Potent Tight Binding Inhibitors.
Pharmaceuticals, 11, 2018
|
|
1ZK2
 
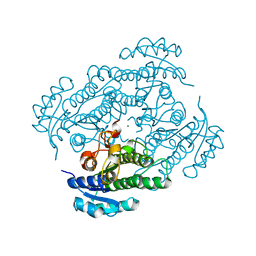 | | Orthorhombic crystal structure of the apo-form of R-specific alcohol dehydrogenase (mutant G37D) from Lactobacillus brevis | | Descriptor: | MAGNESIUM ION, R-specific alcohol dehydrogenase | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Atomic resolution structures of R-specific alcohol dehydrogenase from Lactobacillus brevis provide the structural bases of its substrate and cosubstrate specificity
J.Mol.Biol., 349, 2005
|
|
1ZK0
 
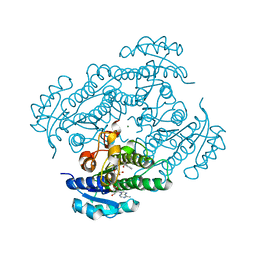 | | Structure of R-specific alcohol dehydrogenase (mutant G37D) from Lactobacillus brevis in complex with phenylethanol and NADH | | Descriptor: | (1R)-1-PHENYLETHANOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
1ZJZ
 
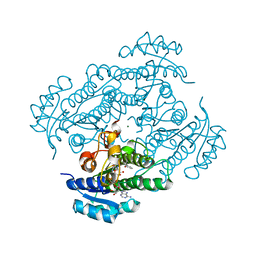 | | Structure of R-specific alcohol dehydrogenase (mutant G37D) from Lactobacillus brevis in complex with phenylethanol and NAD | | Descriptor: | (1R)-1-PHENYLETHANOL, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
1ZK1
 
 | | Structure of R-specific alcohol dehydrogenase (mutant G37D) from Lactobacillus brevis in complex with phenylethanol and NAD | | Descriptor: | 1-PHENYLETHANONE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
1ZK3
 
 | | Triclinic crystal structure of the apo-form of R-specific alcohol dehydrogenase (mutant G37D) from Lactobacillus brevis | | Descriptor: | MAGNESIUM ION, R-specific alcohol dehydrogenase | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
1ZK4
 
 | | Structure of R-specific alcohol dehydrogenase (wildtype) from Lactobacillus brevis in complex with acetophenone and NADP | | Descriptor: | 1-PHENYLETHANONE, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
1ZJY
 
 | | Structure of R-specific alcohol dehydrogenase (mutant G37D) from Lactobacillus brevis in complex with phenylethanol and NADH | | Descriptor: | (1R)-1-PHENYLETHANOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
2JAE
 
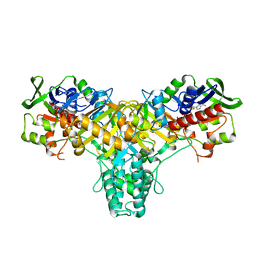 | | The structure of L-amino acid oxidase from Rhodococcus opacus in the unbound state | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE | | Authors: | Faust, A, Niefind, K, Hummel, W, Schomburg, D. | | Deposit date: | 2006-11-27 | | Release date: | 2007-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Structure of a Bacterial L-Amino Acid Oxidase from Rhodococcus Opacus Gives New Evidence for the Hydride Mechanism for Dehydrogenation.
J.Mol.Biol., 367, 2007
|
|
2JB3
 
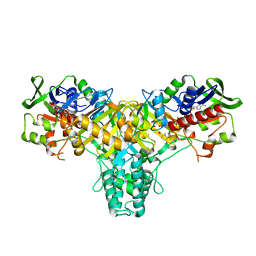 | | The structure of L-amino acid oxidase from Rhodococcus opacus in complex with o-aminobenzoate | | Descriptor: | 2-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE | | Authors: | Faust, A, Niefind, K, hummel, W, Schomburg, D. | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of a Bacterial L-Amino Acid Oxidase from Rhodococcus Opacus Gives New Evidence for the Hydride Mechanism for Dehydrogenation
J.Mol.Biol., 367, 2007
|
|
2JB2
 
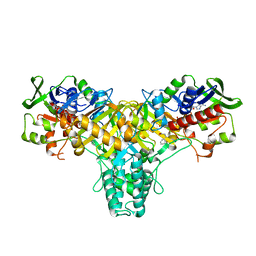 | | The structure of L-amino acid oxidase from Rhodococcus opacus in complex with L-phenylalanine. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE, PHENYLALANINE | | Authors: | Faust, A, Niefind, K, Hummel, W, Schomburg, D. | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Structure of a Bacterial L-Amino Acid Oxidase from Rhodococcus Opacus Gives New Evidence for the Hydride Mechanism for Dehydrogenation
J.Mol.Biol., 367, 2007
|
|
2JB1
 
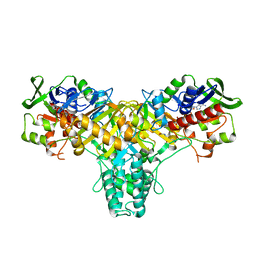 | | The L-amino acid oxidase from Rhodococcus opacus in complex with L- alanine | | Descriptor: | ALANINE, FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE | | Authors: | Faust, A, Niefind, K, Hummel, W, Schomburg, D. | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Structure of a Bacterial L-Amino Acid Oxidase from Rhodococcus Opacus Gives New Evidence for the Hydride Mechanism for Dehydrogenation
J.Mol.Biol., 367, 2007
|
|
4RLL
 
 | | Crystal structure of human CK2alpha in complex with the ATP-competitive inhibitor 4-[(E)-(fluoren-9-ylidenehydrazinylidene)-methyl] benzoate | | Descriptor: | 4-[(E)-(9H-fluoren-9-ylidenehydrazinylidene)methyl]benzoic acid, Casein kinase II subunit alpha, GLYCEROL | | Authors: | Guerra, B, Rasmussen, T.D.L, Schnitzler, A, Jensen, H.H, Boldyreff, B.S, Miyata, Y, Marcussen, N, Niefind, K, Issinger, O.G. | | Deposit date: | 2014-10-17 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Protein kinase CK2 inhibition is associated with the destabilization of HIF-1 alpha in human cancer cells.
Cancer Lett, 356, 2015
|
|
4RLK
 
 | | Crystal structure of Z. mays CK2alpha in complex with the ATP-competitive inhibitor 4-[(E)-(fluoren-9-ylidenehydrazinylidene)-methyl] benzoate | | Descriptor: | 4-[(E)-(9H-fluoren-9-ylidenehydrazinylidene)methyl]benzoic acid, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Guerra, B, Rasmussen, T.D.L, Schnitzler, A, Jensen, H.H, Boldyreff, B.S, Miyata, Y, Marcussen, N, Niefind, K, Issinger, O.G. | | Deposit date: | 2014-10-17 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Protein kinase CK2 inhibition is associated with the destabilization of HIF-1 alpha in human cancer cells.
Cancer Lett, 356, 2015
|
|
1PJK
 
 | | Crystal Structure of a C-terminal deletion mutant of human protein kinase CK2 catalytic subunit | | Descriptor: | CHLORIDE ION, Casein kinase II, alpha chain, ... | | Authors: | Ermakova, I, Boldyreff, B, Issinger, O.-G, Niefind, K. | | Deposit date: | 2003-06-03 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a C-terminal deletion mutant of human protein kinase CK2 catalytic subunit
J.Mol.Biol., 330, 2003
|
|
5OMY
 
 | | HIGH-SALT STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR 4P | | Descriptor: | 4-(3-methylbut-2-enoxy)-5-propan-2-yl-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Hochscherf, J, Lindenblatt, D, Witulski, B, Birus, R, Aichele, D, Marminon, C, Bouaziz, Z, Le Borgne, M, Jose, J, Niefind, K. | | Deposit date: | 2017-08-02 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unexpected Binding Mode of a Potent Indeno[1,2-b]indole-Type Inhibitor of Protein Kinase CK2 Revealed by Complex Structures with the Catalytic Subunit CK2 alpha and Its Paralog CK2 alpha '.
Pharmaceuticals (Basel), 10, 2017
|
|
