1TLQ
 
 | | Crystal structure of protein ypjQ from Bacillus subtilis, Pfam DUF64 | | Descriptor: | CALCIUM ION, Hypothetical protein ypjQ | | Authors: | Kniewel, R, Rajashankar, K.R, Solorzano, V, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-09 | | Release date: | 2004-06-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a Hypothetical Protein from Bacillus subtilis
To be Published
|
|
2FS2
 
 | | Structure of the E. coli PaaI protein from the phyenylacetic acid degradation operon | | Descriptor: | Phenylacetic acid degradation protein paaI, SULFATE ION | | Authors: | Kniewel, R, Buglino, J.A, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-01-20 | | Release date: | 2006-02-07 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, Function, and Mechanism of the Phenylacetate Pathway Hot Dog-fold Thioesterase PaaI
J.Biol.Chem., 281, 2006
|
|
1PUG
 
 | | Structure of E. coli Ybab | | Descriptor: | Hypothetical UPF0133 protein ybaB | | Authors: | Kniewel, R, Buglino, J, Chadna, T, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-24 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of E. coli Ybab
To be Published
|
|
1LA2
 
 | | Structural analysis of Saccharomyces cerevisiae myo-inositol phosphate synthase | | Descriptor: | Myo-inositol-1-phosphate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kniewel, R, Buglino, J.A, Shen, V, Chadna, T, Beckwith, A, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-03-27 | | Release date: | 2002-04-10 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural analysis of Saccharomyces cerevisiae myo-inositol phosphate synthase
J.STRUCT.FUNCT.GENOM., 2, 2002
|
|
1NI3
 
 | |
1P1M
 
 | | Structure of Thermotoga maritima amidohydrolase TM0936 bound to Ni and methionine | | Descriptor: | Hypothetical protein TM0936, METHIONINE, NICKEL (II) ION | | Authors: | Kniewel, R, Buglino, J.A, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-04-12 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the hypothetical protein TM0936 from Thermotoga maritima at
1.5A bound to Ni and methionine
To be Published, 2003
|
|
1P1L
 
 | |
1Q98
 
 | | Structure of a Thiol Peroxidase from Haemophilus influenzae Rd | | Descriptor: | Thiol Peroxidase | | Authors: | Kniewel, R, Buglino, J, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-08-22 | | Release date: | 2003-09-09 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Thiol Peroxidase from Haemophilus influenzae Rd
To be Published
|
|
1PUJ
 
 | | Structure of B. subtilis YlqF GTPase | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, conserved hypothetical protein ylqF | | Authors: | Kniewel, R, Buglino, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-24 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the YlqF GTPase from B. subtilis
To be Published
|
|
1PSU
 
 | | Structure of the E. coli PaaI protein from the phyenylacetic acid degradation operon | | Descriptor: | Phenylacetic acid degradation protein PaaI | | Authors: | Kniewel, R, Buglino, J, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-21 | | Release date: | 2003-07-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, function, and mechanism of the phenylacetate pathway hot dog-fold thioesterase PaaI.
J.Biol.Chem., 281, 2006
|
|
1PUI
 
 | |
1PSW
 
 | | Structure of E. coli ADP-heptose lps heptosyltransferase II | | Descriptor: | ADP-HEPTOSE LPS HEPTOSYLTRANSFERASE II | | Authors: | Kniewel, R, Buglino, J, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-21 | | Release date: | 2003-07-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of E. coli ADP-heptose lps heptosyltransferase II
To be Published
|
|
1PSQ
 
 | | Structure of a probable thiol peroxidase from Streptococcus pneumoniae | | Descriptor: | probable thiol peroxidase | | Authors: | Kniewel, R, Buglino, J, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-21 | | Release date: | 2003-07-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a probable thiol peroxidase from Streptococcus pneumoniae
To be Published
|
|
4LZ6
 
 | | Structure of MATE multidrug transporter DinF-BH | | Descriptor: | BH2163 protein | | Authors: | Lu, M, Radchenko, M, Symersky, J, Nie, R, Guo, Y. | | Deposit date: | 2013-07-31 | | Release date: | 2013-10-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into H(+)-coupled multidrug extrusion by a MATE transporter
Nat.Struct.Mol.Biol., 20, 2013
|
|
4LZ9
 
 | | Structure of MATE multidrug transporter DinF-BH in complex with R6G | | Descriptor: | BH2163 protein, RHODAMINE 6G | | Authors: | Lu, M, Radchenko, M, Symersky, J, Nie, R, Guo, Y. | | Deposit date: | 2013-07-31 | | Release date: | 2013-10-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural insights into H(+)-coupled multidrug extrusion by a MATE transporter
Nat.Struct.Mol.Biol., 20, 2013
|
|
4UXV
 
 | | Cytoplasmic domain of bacterial cell division protein EzrA | | Descriptor: | SEPTATION RING FORMATION REGULATOR EZRA | | Authors: | Cleverley, R.M, Barrett, J.R, Basle, A, Khai-Bui, N, Hewitt, L, Solovyova, A, Xu, Z, Daniela, R.A, Dixon, N.E, Harry, E.J, Oakley, A.J, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.961 Å) | | Cite: | Structure and Function of a Spectrin-Like Regulator of Bacterial Cytokinesis.
Nat.Commun., 5, 2014
|
|
1T35
 
 | | CRYSTAL STRUCTURE OF A HYPOTHETICAL PROTEIN YVDD- A PUTATIVE LYSINE DECARBOXYLASE | | Descriptor: | HYPOTHETICAL PROTEIN YVDD, Putative Lysine Decarboxylase, SULFATE ION | | Authors: | Rajashankar, K.R, Kniewel, R, Solorzano, V, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal Structure of a Hypothetical Protein Yvdd - Putative Lysine Decarboxylase
To be Published
|
|
1SZQ
 
 | | Crystal Structure of 2-methylcitrate dehydratase | | Descriptor: | 2-methylcitrate dehydratase | | Authors: | Rajashankar, K.R, Kniewel, R, Solorzano, V, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-04-06 | | Release date: | 2004-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of 2-methylcitrate dehydratase
To be Published
|
|
1TK9
 
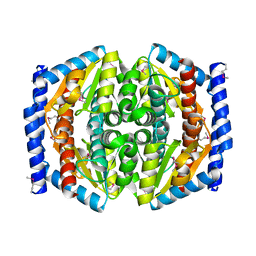 | | Crystal Structure of Phosphoheptose isomerase 1 | | Descriptor: | Phosphoheptose isomerase 1 | | Authors: | Rajashankar, K.R, Solorzano, V, Kniewel, R, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-08 | | Release date: | 2004-06-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of two putative phosphoheptose isomerases.
Proteins, 63, 2006
|
|
1TIK
 
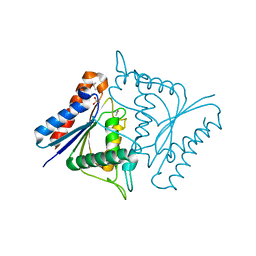 | | CRYSTAL STRUCTURE OF ACYL CARRIER PROTEIN PHOSPHODIESTERASE | | Descriptor: | Acyl carrier protein phosphodiesterase, SULFATE ION | | Authors: | Rajashankar, K.R, Kniewel, R, Solorzano, V, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF ACYL CARRIER PROTEIN PHOSPHODIESTERASE
To be Published
|
|
1TLT
 
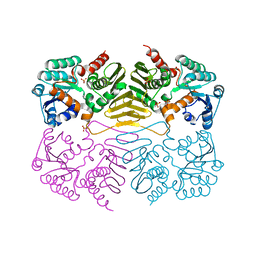 | | Crystal Structure of a Putative Oxidoreductase (VIRULENCE FACTOR mviM HOMOLOG) | | Descriptor: | PUTATIVE OXIDOREDUCTASE (VIRULENCE FACTOR mviM HOMOLOG), SULFATE ION | | Authors: | Rajashankar, K.R, Solorzano, V, Kniewel, R, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-09 | | Release date: | 2004-06-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of a Putative Oxidoreductase (VIRULENCE FACTOR mviM HOMOLOG)
To be Published
|
|
4UY3
 
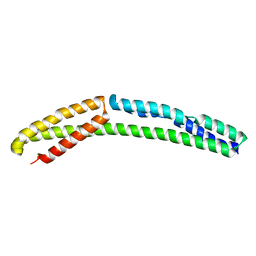 | | Cytoplasmic domain of bacterial cell division protein ezra | | Descriptor: | SEPTATION RING FORMATION REGULATOR EZRA | | Authors: | Cleverley, R.M, Barrett, J.R, Basle, A, Khai-Bui, N, Hewitt, L, Solovyova, A, Xu, Z, Daniela, R.A, Dixon, N.E, Harry, E.J, Oakley, A.J, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Function of a Spectrin-Like Regulator of Bacterial Cytokinesis.
Nat.Commun., 5, 2014
|
|
6G7O
 
 | | Crystal structure of human alkaline ceramidase 3 (ACER3) at 2.7 Angstrom resolution | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Alkaline ceramidase 3,Soluble cytochrome b562, CALCIUM ION, ... | | Authors: | Leyrat, C, Vasiliauskaite-Brooks, I, Healey, R.D, Sounier, R, Grison, C, Hoh, F, Basu, S, Granier, S. | | Deposit date: | 2018-04-06 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a human intramembrane ceramidase explains enzymatic dysfunction found in leukodystrophy.
Nat Commun, 9, 2018
|
|
2XXQ
 
 | | A widespread family of bacterial cell wall assembly proteins | | Descriptor: | (2Z,6Z,10Z,14Z,18Z,22Z,26Z)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl trihydrogen diphosphate, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Marles-Wright, J, Kawai, Y, Emmins, R, Ishikawa, S, Kuwano, M, Heinz, N, Cleverley, R.M, Bui, N.K, Ogasawara, N, Lewis, R.J, Vollmer, W, Daniel, R.A, Errington, J. | | Deposit date: | 2010-11-11 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A Widespread Family of Bacterial Cell Wall Assembly Proteins.
Embo J., 30, 2011
|
|
2YPG
 
 | | Haemagglutinin of 1968 Human H3N2 Virus in Complex with Human Receptor Analogue LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, J, Xiong, X, Haire, L.F, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | Deposit date: | 2012-10-30 | | Release date: | 2012-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
