2OG9
 
 | |
2OGK
 
 | |
3QJK
 
 | |
2NWU
 
 | |
1SR6
 
 | | Structure of nucleotide-free scallop myosin S1 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Myosin essential light chain, ... | | Authors: | Risal, D, Gourinath, S, Himmel, D.M, Szent-Gyorgyi, A.G, Cohen, C. | | Deposit date: | 2004-03-22 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Myosin subfragment 1 structures reveal a partially bound nucleotide and a complex salt bridge that helps couple nucleotide and actin binding.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
2OPJ
 
 | |
2OOD
 
 | |
2P0L
 
 | |
2OY9
 
 | |
2P9B
 
 | |
2PBZ
 
 | |
1YV9
 
 | |
1ZL6
 
 | | Crystal structure of Tyr350Ala mutant of Clostridium botulinum neurotoxin E catalytic domain | | Descriptor: | SULFATE ION, ZINC ION, botulinum neurotoxin type E | | Authors: | Agarwal, R, Binz, T, Swaminathan, S. | | Deposit date: | 2005-05-05 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
1YVG
 
 | | Structural analysis of the catalytic domain of tetanus neurotoxin | | Descriptor: | Tetanus toxin, light chain, ZINC ION | | Authors: | Rao, K.N, Kumaran, D, Binz, T, Swaminathan, S. | | Deposit date: | 2005-02-15 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of the catalytic domain of tetanus neurotoxin.
Toxicon, 45, 2005
|
|
1ZKX
 
 | | Crystal structure of Glu158Ala/Thr159Ala/Asn160Ala- a triple mutant of Clostridium botulinum neurotoxin E catalytic domain | | Descriptor: | CHLORIDE ION, ZINC ION, botulinum neurotoxin type E | | Authors: | Agarwal, R, Binz, T, Swaminathan, S. | | Deposit date: | 2005-05-04 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
2A97
 
 | |
3LF0
 
 | | Crystal structure of the ATP bound Mycobacterium tuberculosis nitrogen regulatory PII protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Nitrogen regulatory protein P-II | | Authors: | Shetty, N.D, Palaninathan, S.K, Reddy, M.C.M, Owen, J.L, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-01-15 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the apo and ATP bound Mycobacterium tuberculosis nitrogen regulatory PII protein.
Protein Sci., 19, 2010
|
|
3PRK
 
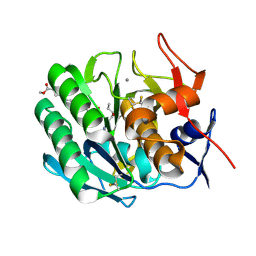 | | INHIBITION OF PROTEINASE K BY METHOXYSUCCINYL-ALA-ALA-PRO-ALA-CHLOROMETHYL KETONE. AN X-RAY STUDY AT 2.2-ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, METHOXYSUCCINYL-ALA-ALA-PRO-ALA-CHLOROMETHYL KETONE, PROTEINASE K | | Authors: | Wolf, W.M, Bajorath, J, Mueller, A, Raghunathan, S, Singh, T.P, Hinrichs, W, Saenger, W. | | Deposit date: | 1991-08-07 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibition of proteinase K by methoxysuccinyl-Ala-Ala-Pro-Ala-chloromethyl ketone. An x-ray study at 2.2-A resolution.
J.Biol.Chem., 266, 1991
|
|
3DEB
 
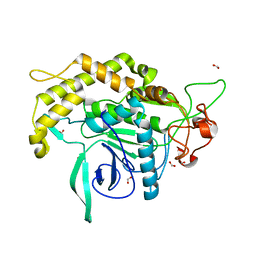 | |
1ZL5
 
 | | Crystal structure of Glu335Gln mutant of Clostridium botulinum neurotoxin E catalytic domain | | Descriptor: | CHLORIDE ION, botulinum neurotoxin type E | | Authors: | Agarwal, R, Binz, T, Swaminathan, S. | | Deposit date: | 2005-05-05 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
1TH2
 
 | | crystal structure of NADPH depleted bovine liver catalase complexed with azide | | Descriptor: | AZIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugadev, R, Balasundaresan, D, Ponnuswamy, M.N, Kumaran, D, Swaminathan, S, Sekar, K. | | Deposit date: | 2004-06-01 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of bovine liver catalase
TO BE PUBLISHED
|
|
1ZN3
 
 | | Crystal structure of Glu335Ala mutant of Clostridium botulinum neurotoxin type E | | Descriptor: | CHLORIDE ION, ZINC ION, botulinum neurotoxin type E | | Authors: | Agarwal, R, Binz, T, Swaminathan, S. | | Deposit date: | 2005-05-11 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
1TH3
 
 | | Crystal structure of NADPH depleted bovine live catalase complexed with cyanide | | Descriptor: | CYANIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugadev, R, Balasundaresan, D, Ponnuswamy, M.N, Kumaran, D, Swaminathan, S, Sekar, K. | | Deposit date: | 2004-06-01 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of bovine liver catalase
TO BE PUBLISHED
|
|
2A8A
 
 | |
1ZKW
 
 | | Crystal structure of Arg347Ala mutant of botulinum neurotoxin E catalytic domain | | Descriptor: | CHLORIDE ION, ZINC ION, botulinum neurotoxin type E | | Authors: | Agarwal, R, Binz, T, Swaminathan, S. | | Deposit date: | 2005-05-04 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
