9UGI
 
 | |
9L9R
 
 | |
9LMK
 
 | |
9LML
 
 | |
9LPU
 
 | | Crystal structure of tKeima at pH 4.0 | | Descriptor: | Large stokes shift fluorescent protein | | Authors: | Nam, K.H. | | Deposit date: | 2025-01-26 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | pH-Induced Conformational Change of the Chromophore of the Large Stokes Shift Fluorescent Protein tKeima.
Molecules, 30, 2025
|
|
8GMV
 
 | | Crystal structure of lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Nam, K.H. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of lysozyme
To Be Published
|
|
8GMW
 
 | | Crystal structure of lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Nam, K.H. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of lysozyme
To Be Published
|
|
8YUD
 
 | |
8YYO
 
 | |
8YJI
 
 | |
8YYN
 
 | |
8YJJ
 
 | |
8YPX
 
 | |
9K73
 
 | | Crystal structure of TsaBgl | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, beta-glucosidase | | Authors: | Nam, K.H. | | Deposit date: | 2024-10-23 | | Release date: | 2024-11-06 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Application of Serial Crystallography for Merging Incomplete Macromolecular Crystallography Datasets.
Crystals, 14, 2024
|
|
9K9Z
 
 | |
9KA0
 
 | |
9K72
 
 | | Crystal structure of TsaBgl using merged datasets | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, beta-glucosidase | | Authors: | Nam, K.H. | | Deposit date: | 2024-10-23 | | Release date: | 2024-11-06 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Application of Serial Crystallography for Merging Incomplete Macromolecular Crystallography Datasets.
Crystals, 14, 2024
|
|
8ZM4
 
 | |
8ZM6
 
 | |
8ZM5
 
 | |
8IH0
 
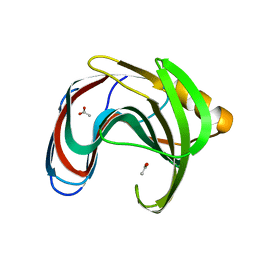 | | Crystal structure of GH11 from Thermoanaerobacterium saccharolyticum | | Descriptor: | ACETATE ION, Endo-1,4-beta-xylanase | | Authors: | Nam, K.H. | | Deposit date: | 2023-02-22 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization and structural analysis of the endo-1,4-beta-xylanase GH11 from the hemicellulose-degrading Thermoanaerobacterium saccharolyticum useful for lignocellulose saccharification.
Sci Rep, 13, 2023
|
|
8IH1
 
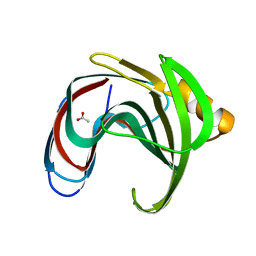 | |
3L1H
 
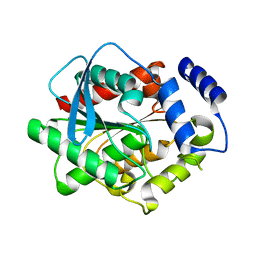 | | Crystal structure of EstE5, was soaked by FeCl3 | | Descriptor: | Esterase/lipase | | Authors: | Nam, K.H, Hwang, K.Y. | | Deposit date: | 2009-12-11 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the noninvasive inhibition of HSL-homolog EstE5 by organic solvents and metal ions
To be Published
|
|
3L1I
 
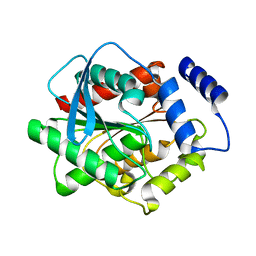 | | Crystal structure of EstE5, was soaked by CuSO4 | | Descriptor: | Esterase/lipase | | Authors: | Nam, K.H, Hwang, K.Y. | | Deposit date: | 2009-12-11 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the noninvasive inhibition of HSL-homolog EstE5 by organic solvents and metal ions
To be Published
|
|
3L1J
 
 | | Crystal structure of EstE5, was soaked by ZnSO4 | | Descriptor: | Esterase/lipase | | Authors: | Nam, K.H, Hwang, K.Y. | | Deposit date: | 2009-12-11 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the noninvasive inhibition of HSL-homolog EstE5 by organic solvents and metal ions
To be Published
|
|
