7DFJ
 
 | |
7DFK
 
 | |
7DIG
 
 | |
7E25
 
 | | Crystal structure of human FABP7 complexed with palmitic acid | | Descriptor: | Fatty acid-binding protein, brain, GLYCEROL, ... | | Authors: | Nam, K.H. | | Deposit date: | 2021-02-04 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of human brain-type fatty acid-binding protein FABP7 complexed with palmitic acid.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7E5J
 
 | | Crystal structure of beta-glucosidase from Thermoanaerobacterium saccharolyticum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase, SODIUM ION | | Authors: | Nam, K.H. | | Deposit date: | 2021-02-18 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Biochemical and Structural Analysis of a Glucose-Tolerant beta-Glucosidase from the Hemicellulose-Degrading Thermoanaerobacterium saccharolyticum.
Molecules, 27, 2022
|
|
7DTF
 
 | |
7E02
 
 | |
7E03
 
 | |
7DTB
 
 | |
7CVK
 
 | |
7CVJ
 
 | |
7CVL
 
 | |
7CJP
 
 | | Crystal structure of metal-free state of glucose isomerase | | Descriptor: | 1,2-ETHANEDIOL, Xylose isomerase | | Authors: | Nam, K.H. | | Deposit date: | 2020-07-12 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the metal-free state of glucose isomerase reveals its minimal open configuration for metal binding.
Biochem.Biophys.Res.Commun., 547, 2021
|
|
7CJO
 
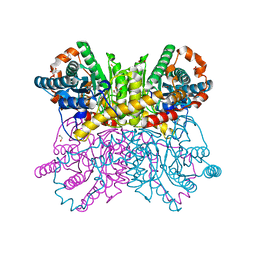 | | Crystal structure of metal-bound state of glucose isomerase | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Xylose isomerase | | Authors: | Nam, K.H. | | Deposit date: | 2020-07-12 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the metal-free state of glucose isomerase reveals its minimal open configuration for metal binding.
Biochem.Biophys.Res.Commun., 547, 2021
|
|
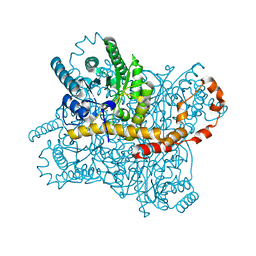
7CJZ
 
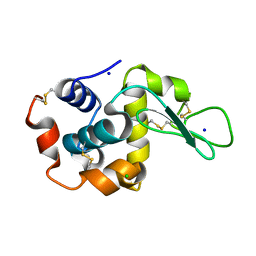 | |
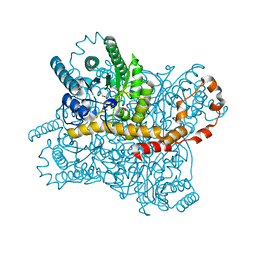
7CK0
 
 | |
5ZYC
 
 | | Crystal Structure of Glucose Isomerase Soaked with Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Nam, K.H. | | Deposit date: | 2018-05-24 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of substrate recognition by glucose isomerase in Mn2+binding mode at M2 site in S. rubiginosus
Biochem. Biophys. Res. Commun., 503, 2018
|
|
5ZYE
 
 | | Crystal Structure of Glucose Isomerase Soaked with Mn2+ and Glucose | | Descriptor: | MANGANESE (II) ION, Xylose isomerase, alpha-D-glucopyranose | | Authors: | Nam, K.H. | | Deposit date: | 2018-05-24 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural analysis of substrate recognition by glucose isomerase in Mn2+binding mode at M2 site in S. rubiginosus
Biochem. Biophys. Res. Commun., 503, 2018
|
|
5ZYD
 
 | | Crystal Structure of Glucose Isomerase Soaked with Glucose | | Descriptor: | ACETATE ION, MAGNESIUM ION, Xylose isomerase | | Authors: | Nam, K.H. | | Deposit date: | 2018-05-24 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural analysis of substrate recognition by glucose isomerase in Mn2+binding mode at M2 site in S. rubiginosus
Biochem. Biophys. Res. Commun., 503, 2018
|
|
8D9H
 
 | | gRAMP-TPR-CHAT match PFS target RNA(Craspase) | | Descriptor: | CHAT domain protein, PHOSPHATE ION, RAMP superfamily protein, ... | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
8D9E
 
 | | gRAMP-match PFS target | | Descriptor: | RAMP superfamily protein, RNA (36-MER), RNA (5'-R(P*UP*CP*CP*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP*GP*UP*A)-3'), ... | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
8D97
 
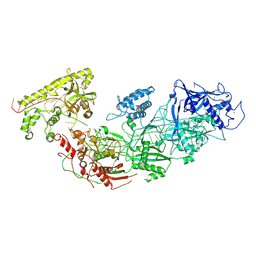 | | Apo gRAMP | | Descriptor: | RAMP superfamily protein, RNA (42-MER), ZINC ION | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
8D9F
 
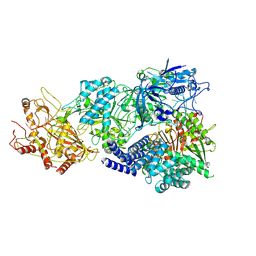 | | gRAMP-TPR-CHAT (Craspase) | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
8D9G
 
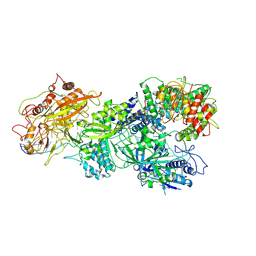 | | gRAMP-TPR-CHAT Non match PFS target RNA(Craspase) | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (36-MER), ... | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
8D9I
 
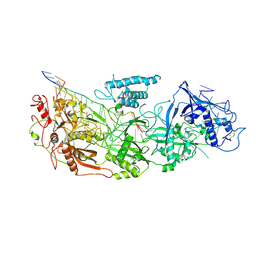 | | gRAMP non-matching PFS-with Mg | | Descriptor: | RAMP superfamily protein, RNA (35-MER), RNA (5'-R(P*UP*CP*CP*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP*A)-3'), ... | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
