4MMI
 
 | | Crystal structure of heparan sulfate lyase HepC mutant from Pedobacter heparinus | | Descriptor: | CALCIUM ION, Heparinase III protein | | Authors: | Maruyama, Y, Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2013-09-09 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Pedobacter heparinus Heparin Lyase Hep III with the Active Site in a Deep Cleft
Biochemistry, 53, 2014
|
|
7WDO
 
 | | Crystal structures of MeBglD2 in complex with various saccharides | | Descriptor: | Beta-glucosidase, SULFATE ION, beta-D-glucopyranose, ... | | Authors: | Watanabe, M, Matsuzawa, T, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-12-22 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of metagenomic beta-glycosidase MeBglD2 in complex with various saccharides
Appl.Microbiol.Biotechnol., 106, 2022
|
|
7WDP
 
 | | Crystal structures of MeBglD2 in complex with various saccharides | | Descriptor: | Beta-glucosidase, SULFATE ION, alpha-D-glucopyranose, ... | | Authors: | Watanabe, M, Matsuzawa, T, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-12-22 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of metagenomic beta-glycosidase MeBglD2 in complex with various saccharides
Appl.Microbiol.Biotechnol., 106, 2022
|
|
7WDR
 
 | | Crystal structures of MeBglD2 in complex with various saccharides | | Descriptor: | 4-nitrophenyl beta-D-glucopyranoside, Beta-glucosidase, SULFATE ION | | Authors: | Watanabe, M, Matsuzawa, T, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-12-22 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of metagenomic beta-glycosidase MeBglD2 in complex with various saccharides
Appl.Microbiol.Biotechnol., 106, 2022
|
|
7WDS
 
 | | Crystal structures of MeBglD2 in complex with various saccharides | | Descriptor: | Beta-glucosidase, SULFATE ION, beta-D-xylopyranose | | Authors: | Watanabe, M, Matsuzawa, T, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-12-22 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of metagenomic beta-glycosidase MeBglD2 in complex with various saccharides
Appl.Microbiol.Biotechnol., 106, 2022
|
|
7WDV
 
 | | Crystal structures of MeBglD2 in complex with various saccharides | | Descriptor: | Beta-glucosidase, SULFATE ION, beta-D-glucopyranose, ... | | Authors: | Watanabe, M, Matsuzawa, T, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-12-22 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.812 Å) | | Cite: | Crystal structure of metagenomic beta-glycosidase MeBglD2 in complex with various saccharides
Appl.Microbiol.Biotechnol., 106, 2022
|
|
7WDN
 
 | | Crystal structures of MeBglD2 in complex with various saccharides | | Descriptor: | alpha-D-glucopyranose, beta-glucosidase | | Authors: | Watanabe, M, Matsuzawa, T, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-12-22 | | Release date: | 2023-01-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of metagenomic beta-glycosidase MeBglD2 in complex with various saccharides.
Appl.Microbiol.Biotechnol., 106, 2022
|
|
7EAP
 
 | | Crystal structure of IpeA-XXXG complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Matsuzawa, T, Watanabe, M, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural basis for the catalytic mechanism of the glycoside hydrolase family 3 isoprimeverose-producing oligoxyloglucan hydrolase from Aspergillus oryzae.
Febs Lett., 596, 2022
|
|
7ETQ
 
 | | Crystal structure of Pro-Met-Leu-Leu | | Descriptor: | Pro-Met-Leu-Leu | | Authors: | Kurumida, Y, Ikeda, K, Nakamichi, Y, Hirano, A, Kobayashi, K, Saito, Y, Kameda, T. | | Deposit date: | 2021-05-13 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystal structure of Pro-Met-Leu-Leu
To Be Published
|
|
7ETP
 
 | | Crystal structure of Pro-Phe-Leu-Phe | | Descriptor: | Pro-Phe-Leu-Phe | | Authors: | Kurumida, Y, Ikeda, K, Nakamichi, Y, Hirano, A, Kobayashi, K, Saito, Y, Kameda, T. | | Deposit date: | 2021-05-13 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.09488 Å) | | Cite: | Crystal structure of Pro-Phe-Leu-Phe
To Be Published
|
|
7ETN
 
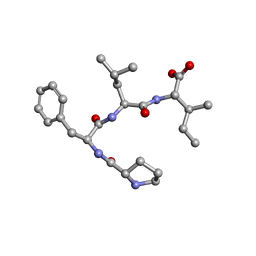 | | Crystal structure of Pro-Phe-Leu-Ile | | Descriptor: | PRO-PHE-LEU-ILE | | Authors: | Kurumida, Y, Ikeda, K, Nakamichi, Y, Hirano, A, Kobayashi, K, Saito, Y, Kameda, T. | | Deposit date: | 2021-05-13 | | Release date: | 2022-05-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (0.82 Å) | | Cite: | Crystal structure of Pro-Phe-Leu-Ile
To Be Published
|
|
5ZN7
 
 | |
5YOT
 
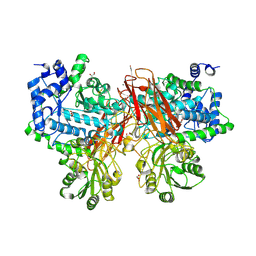 | | Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Matsuzawa, T, Watanabe, M, Nakamichi, Y, Yaoi, K. | | Deposit date: | 2017-10-31 | | Release date: | 2018-11-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure and substrate recognition mechanism of Aspergillus oryzae isoprimeverose-producing enzyme.
J.Struct.Biol., 205, 2019
|
|
5YQS
 
 | | Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yaoi, K, Matsuzawa, T, Watanabe, M, Nakamichi, Y. | | Deposit date: | 2017-11-07 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and substrate recognition mechanism of Aspergillus oryzae isoprimeverose-producing enzyme.
J.Struct.Biol., 205, 2019
|
|
5ZN6
 
 | |
