5FLL
 
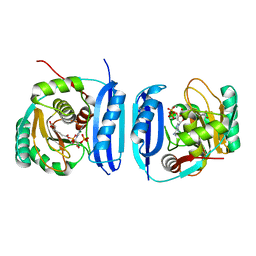 | | Crystal structure of the 6-carboxyhexanoate-CoA ligase (BioW) from Bacillus subtilis in complex with a Pimeloyl-adenylate | | Descriptor: | 6-CARBOXYHEXANOATE-COA LIGASE, MAGNESIUM ION, PIMELOYL-AMP, ... | | Authors: | Moynie, L, Wang, M, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2015-10-26 | | Release date: | 2016-11-16 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Using the pimeloyl-CoA synthetase adenylation fold to synthesize fatty acid thioesters.
Nat. Chem. Biol., 13, 2017
|
|
5FLG
 
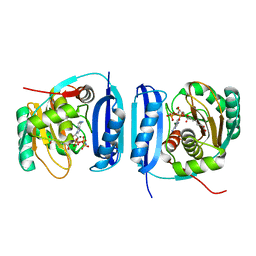 | | Crystal structure of the 6-carboxyhexanoate-CoA ligase (BioW)from Bacillus subtilis in complex with AMPPNP | | Descriptor: | 6-CARBOXYHEXANOATE--COA LIGASE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Moynie, L, Wang, M, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2015-10-26 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Using the pimeloyl-CoA synthetase adenylation fold to synthesize fatty acid thioesters.
Nat. Chem. Biol., 13, 2017
|
|
1I3H
 
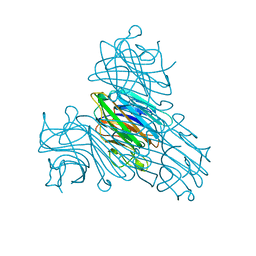 | | CONCANAVALIN A-DIMANNOSE STRUCTURE | | Descriptor: | CALCIUM ION, Concanavalin-A, MANGANESE (II) ION, ... | | Authors: | Sanders, D.A.R, Moothoo, D.N, Raftery, J, Howard, A.J, Helliwell, J.R, Naismith, J.H. | | Deposit date: | 2001-02-15 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A resolution structure of the Con A-dimannose complex.
J.Mol.Biol., 310, 2001
|
|
1KEP
 
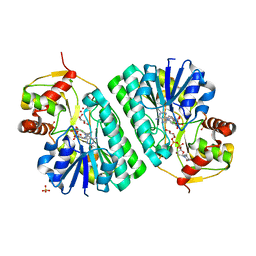 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Streptococcus suis with dTDP-xylose bound | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, THYMIDINE-5'-DIPHOSPHO-BETA-D-XYLOSE, ... | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-16 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
1RQR
 
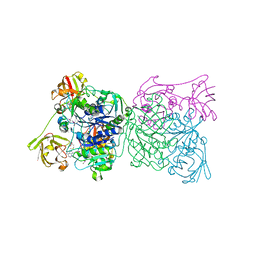 | | Crystal structure and mechanism of a bacterial fluorinating enzyme, product complex | | Descriptor: | 5'-FLUORO-5'-DEOXYADENOSINE, 5'-fluoro-5'-deoxyadenosine synthase, METHIONINE | | Authors: | Dong, C, Huang, F, Deng, H, Schaffrath, C, Spencer, J.B, O'Hagan, D, Naismith, J.H. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2022-06-15 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure and mechanism of a bacterial fluorinating enzyme
Nature, 427, 2004
|
|
1KER
 
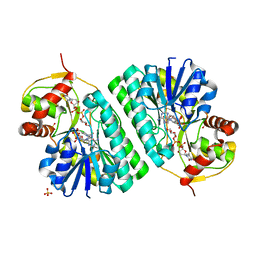 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Streptococcus suis with dTDP-D-glucose bound | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-17 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
5FM0
 
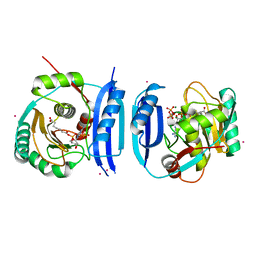 | | Crystal structure of the 6-carboxyhexanoate-CoA ligase (BioW)from Bacillus subtilis (PtCl4 derivative) | | Descriptor: | 6-CARBOXYHEXANOATE--COA LIGASE, MAGNESIUM ION, PIMELOYL-AMP, ... | | Authors: | Moynie, L, Wang, M, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2015-10-29 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Using the pimeloyl-CoA synthetase adenylation fold to synthesize fatty acid thioesters.
Nat. Chem. Biol., 13, 2017
|
|
1OI6
 
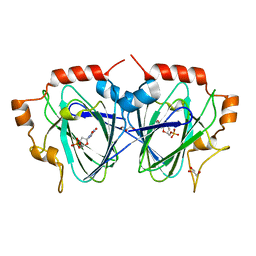 | | Structure determination of the TMP-complex of EvaD | | Descriptor: | GLYCEROL, PCZA361.16, THYMIDINE-5'-PHOSPHATE | | Authors: | Merkel, A.B, Naismith, J.H. | | Deposit date: | 2003-06-09 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Position of a Key Tyrosine in Dtdp-4-Keto-6-Deoxy-D-Glucose-5-Epimerase (Evad) Alters the Substrate Profile for This Rmlc-Like Enzyme
J.Biol.Chem., 279, 2004
|
|
1KEU
 
 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Salmonella enterica serovar Typhimurium with dTDP-D-glucose bound | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, dTDP-D-glucose 4,6-dehydratase | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-17 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
6I2J
 
 | | Crystal structure of the ferric enterobactin receptor mutant (Q482A) from Pseudomonas aeruginosa (PfeA) in complex with enterobactin | | Descriptor: | FE (III) ION, Ferric enterobactin receptor, N,N',N''-[(3S,7S,11S)-2,6,10-trioxo-1,5,9-trioxacyclododecane-3,7,11-triyl]tris(2,3-dihydroxybenzamide) | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2018-11-01 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The complex of ferric-enterobactin with its transporter from Pseudomonas aeruginosa suggests a two-site model.
Nat Commun, 10, 2019
|
|
1KET
 
 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Streptococcus suis with thymidine diphosphate bound | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, THYMIDINE-5'-DIPHOSPHATE, dTDP-D-glucose 4,6-dehydratase | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-17 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
6H7F
 
 | | Crystal structure of BauA, the Ferric preacinetobactin receptor from Acinetobacter baumannii in complex with Fe3+-Preacinetobactin-acinetobactin | | Descriptor: | (4~{S},5~{R})-2-[2,3-bis(oxidanyl)phenyl]-~{N}-[2-(1~{H}-imidazol-4-yl)ethyl]-5-methyl-~{N}-oxidanyl-4,5-dihydro-1,3-oxazole-4-carboxamide, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, ... | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2018-07-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Preacinetobactin not acinetobactin is essential for iron uptake by the BauA transporter of the pathogenAcinetobacter baumannii.
Elife, 7, 2018
|
|
5LQ4
 
 | | The Structure of ThcOx, the First Oxidase Protein from the Cyanobactin Pathways | | Descriptor: | CyaGox, FLAVIN MONONUCLEOTIDE | | Authors: | Bent, A.F, Wagner, A, Naismith, J.H. | | Deposit date: | 2016-08-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the cyanobactin oxidase ThcOx from Cyanothece sp. PCC 7425, the first structure to be solved at Diamond Light Source beamline I23 by means of S-SAD.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6H7V
 
 | |
5FUH
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Tran, F, Westwood, N.J, Naismith, J.H. | | Deposit date: | 2016-01-27 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Allosteric Competitive Inhibitors of the Glucose-1-Phosphate Thymidylyltransferase (Rmla) from Pseudomonas Aeruginosa.
To be Published
|
|
6HCP
 
 | | Crystal structure of BauA, the Ferric preacinetobactin receptor from Acinetobacter baumannii | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, BauA, ... | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Preacinetobactin not acinetobactin is essential for iron uptake by the BauA transporter of the pathogenAcinetobacter baumannii.
Elife, 7, 2018
|
|
5FU0
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Tran, F, Westwood, N.J, Naismith, J.H. | | Deposit date: | 2016-01-19 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric Competitive Inhibitors of the Glucose-1-Phosphate Thymidylyltransferase (Rmla) from Pseudomonas Aeruginosa.
To be Published
|
|
6Q5E
 
 | | Crystal structure of the ferric enterobactin receptor from Pseudomonas aeruginosa (PfeA) in complex with enterobactin | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, FE (III) ION, Ferric enterobactin receptor, ... | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2018-12-07 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The complex of ferric-enterobactin with its transporter from Pseudomonas aeruginosa suggests a two-site model.
Nat Commun, 10, 2019
|
|
1O7I
 
 | |
5FU8
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Tran, F, Westwood, N.J, Naismith, J.H. | | Deposit date: | 2016-01-21 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Allosteric Competitive Inhibitors of the Glucose-1-Phosphate Thymidylyltransferase (Rmla) from Pseudomonas Aeruginosa.
To be Published
|
|
5FYE
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Tran, F, Westwood, N.J, Naismith, J.H. | | Deposit date: | 2016-03-07 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric Competitive Inhibitors of the Glucose-1-Phosphate Thymidylyltransferase (Rmla) from Pseudomonas Aeruginosa.
To be Published
|
|
5FTV
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Tran, F, Westwood, N.J, Naismith, J.H. | | Deposit date: | 2016-01-18 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.213 Å) | | Cite: | Allosteric Competitive Inhibitors of the Glucose-1-Phosphate Thymidylyltransferase (Rmla) from Pseudomonas Aeruginosa.
To be Published
|
|
6QZY
 
 | | full length OphA V406P in complex with SAH | | Descriptor: | ASN-GLY-PHE-PRO-TRP-MVA-ILE-MVA-VAL-GLY-PRO-ILE-GLY, MAGNESIUM ION, Peptide N-methyltransferase, ... | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Substrate Plasticity of a Fungal Peptide alpha-N-Methyltransferase.
Acs Chem.Biol., 15, 2020
|
|
6QZZ
 
 | | full length OphA V404E in complex with SAH | | Descriptor: | Peptide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Plasticity of a Fungal Peptide alpha-N-Methyltransferase.
Acs Chem.Biol., 15, 2020
|
|
6R00
 
 | | OphA DeltaC6 V404F complex with SAH | | Descriptor: | PHE-PRO-TRP-MVA-ILE-MVA-PHE-GLY-VAL-ILE-GLY-VAL-ILE-GLY, Peptide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Substrate Plasticity of a Fungal Peptide alpha-N-Methyltransferase.
Acs Chem.Biol., 15, 2020
|
|
