2ZEY
 
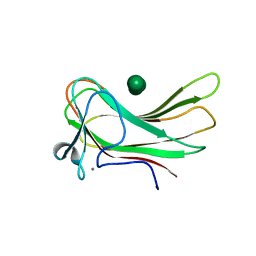 | | Family 16 carbohydrate binding module | | 分子名称: | CALCIUM ION, S-layer associated multidomain endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose, ... | | 著者 | Nair, S.K, Bae, B. | | 登録日 | 2007-12-18 | | 公開日 | 2008-03-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Molecular Basis for the Selectivity and Specificity of Ligand Recognition by the Family 16 Carbohydrate-binding Modules from Thermoanaerobacterium polysaccharolyticum ManA
J.Biol.Chem., 283, 2008
|
|
4NU6
 
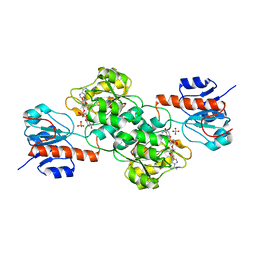 | | Crystal Structure of PTDH R301K | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Phosphonate dehydrogenase, SULFATE ION | | 著者 | Nair, S.K, Chekan, J.R. | | 登録日 | 2013-12-03 | | 公開日 | 2014-03-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Chemical rescue and inhibition studies to determine the role of arg301 in phosphite dehydrogenase.
Plos One, 9, 2014
|
|
4NU5
 
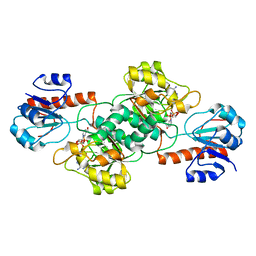 | | Crystal Structure of PTDH R301A | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Phosphonate dehydrogenase | | 著者 | Nair, S.K, Chekan, J.R. | | 登録日 | 2013-12-03 | | 公開日 | 2014-03-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Chemical rescue and inhibition studies to determine the role of arg301 in phosphite dehydrogenase.
Plos One, 9, 2014
|
|
4OJZ
 
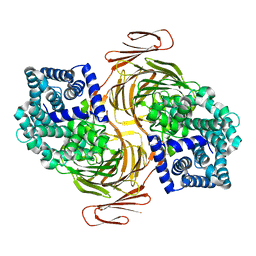 | |
4OK4
 
 | | Crystal Structure of Alg17c Mutant H202L | | 分子名称: | Putative alginate lyase, ZINC ION | | 著者 | Nair, S.K, Park, D.S. | | 登録日 | 2014-01-21 | | 公開日 | 2014-01-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of a PL17 Family Alginate Lyase Demonstrates Functional Similarities among Exotype Depolymerases.
J.Biol.Chem., 289, 2014
|
|
4I3V
 
 | |
4I3U
 
 | |
4I3W
 
 | |
4I3T
 
 | |
4I3X
 
 | |
4H6V
 
 | |
4H6X
 
 | |
4H6W
 
 | |
3SJM
 
 | |
3R9E
 
 | |
3R95
 
 | |
3R9F
 
 | |
3R96
 
 | |
3R9G
 
 | |
3G7D
 
 | | Native PhpD with Cadmium Atoms | | 分子名称: | CADMIUM ION, PhpD | | 著者 | Nair, S.K. | | 登録日 | 2009-02-09 | | 公開日 | 2009-06-09 | | 最終更新日 | 2012-03-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | An unusual carbon-carbon bond cleavage reaction during phosphinothricin biosynthesis.
Nature, 459, 2009
|
|
7QOS
 
 | |
5DLY
 
 | |
5DM1
 
 | |
3UQ4
 
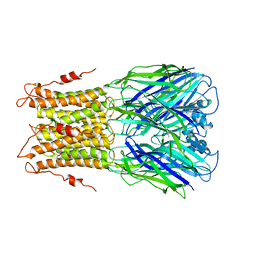 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) mutant F247L (F16L) | | 分子名称: | Gamma-aminobutyric-acid receptor subunit beta-1, SODIUM ION | | 著者 | Gonzalez-Gutierrez, G, Lukk, T, Agarwal, V, Papke, D, Nair, S.K, Grosman, C. | | 登録日 | 2011-11-19 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Mutations that stabilize the open state of the Erwinia chrisanthemi ligand-gated ion channel fail to change the conformation of the pore domain in crystals.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UQ5
 
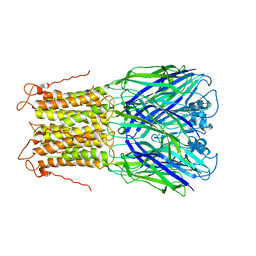 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) mutant L240A F247L (L9A F16L) in the presence of 10 mM cysteamine | | 分子名称: | Gamma-aminobutyric-acid receptor subunit beta-1, SODIUM ION | | 著者 | Gonzalez-Gutierrez, G, Lukk, T, Agarwal, V, Papke, D, Nair, S.K, Grosman, C. | | 登録日 | 2011-11-19 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (4.2 Å) | | 主引用文献 | Mutations that stabilize the open state of the Erwinia chrisanthemi ligand-gated ion channel fail to change the conformation of the pore domain in crystals.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
