1UHN
 
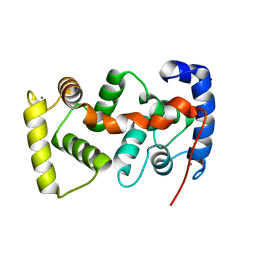 | | The crystal structure of the calcium binding protein AtCBL2 from Arabidopsis thaliana | | Descriptor: | CALCIUM ION, calcineurin B-like protein 2 | | Authors: | Nagae, M, Nozawa, A, Koizumi, N, Sano, H, Hashimoto, H, Sato, M, Shimizu, T. | | Deposit date: | 2003-07-07 | | Release date: | 2003-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of the Novel Calcium-binding Protein AtCBL2 from Arabidopsis thaliana
J.Biol.Chem., 278, 2003
|
|
5XFI
 
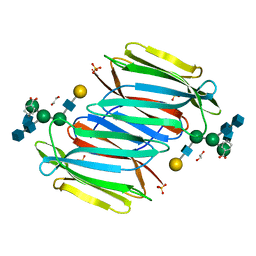 | |
5XFH
 
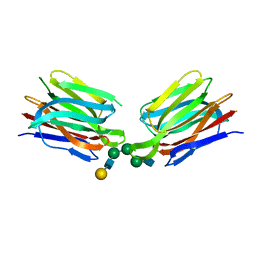 | | Crystal structure of Orysata lectin in complex with biantennary N-glycan | | Descriptor: | Salt stress-induced protein, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2017-04-10 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Distinct roles for each N-glycan branch interacting with mannose-binding type Jacalin-related lectins Orysata and Calsepa.
Glycobiology, 27, 2017
|
|
7F5K
 
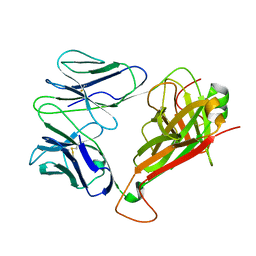 | | Crystal structure of TCR4-1 ectodomain | | Descriptor: | T cell receptor alpha chain, T cell receptor beta chain | | Authors: | Nagae, M, Yamasaki, S. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.0000298 Å) | | Cite: | Symbiotic bacteria-dependent expansion of MR1-reactive T cells causes autoimmunity in the absence of Bcl11b.
Nat Commun, 13, 2022
|
|
2EAB
 
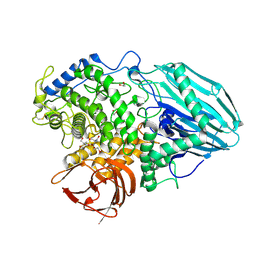 | | Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum (apo form) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-fucosidase, ... | | Authors: | Nagae, M, Tsuchiya, A, Katayama, T, Yamamoto, K, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural basis on the catalytic reaction mechanism of novel 1,2-alpha-L-fucosidase (AFCA) from Bifidobacterium bifidum
J.Biol.Chem., 282, 2007
|
|
2EAC
 
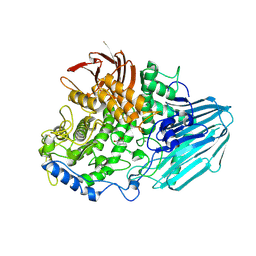 | | Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with deoxyfuconojirimycin | | Descriptor: | (2S,3R,4S,5R)-2-METHYLPIPERIDINE-3,4,5-TRIOL, Alpha-fucosidase, CALCIUM ION | | Authors: | Nagae, M, Tsuchiya, A, Katayama, T, Yamamoto, K, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-04-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis on the catalytic reaction mechanism of novel 1,2-alpha-L-fucosidase (AFCA) from Bifidobacterium bifidum
J.Biol.Chem., 282, 2007
|
|
2EAL
 
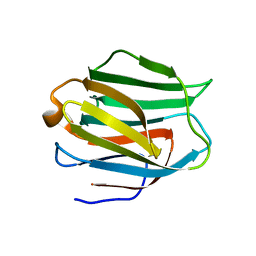 | | Crystal structure of human galectin-9 N-terminal CRD in complex with Forssman pentasaccharide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-beta-D-galactopyranose, Galectin-9 | | Authors: | Nagae, M, Nakamura-Tsuruta, S, Nishi, N, Nakamura, T, Hirabayashi, J, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-09-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of the human galectin-9 N-terminal carbohydrate recognition domain reveals unexpected properties that differ from the mouse orthologue.
J.Mol.Biol., 375, 2008
|
|
2EAK
 
 | | Crystal structure of human galectin-9 N-terminal CRD in complex with lactose | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, GLYCEROL, Galectin-9, ... | | Authors: | Nagae, M, Nakamura-Tsuruta, S, Nishi, N, Nakamura, T, Hirabayashi, J, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-09-25 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural analysis of the human galectin-9 N-terminal carbohydrate recognition domain reveals unexpected properties that differ from the mouse orthologue.
J.Mol.Biol., 375, 2008
|
|
5GZ9
 
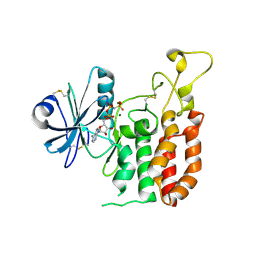 | | Crystal structure of catalytic domain of Protein O-mannosyl Kinase in complexes with AMP-PNP, Magnesium ions and glycopeptide | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein O-mannose kinase, ... | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2016-09-27 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3D structural analysis of protein O-mannosyl kinase, POMK, a causative gene product of dystroglycanopathy.
Genes Cells, 22, 2017
|
|
5GZ8
 
 | |
5GU5
 
 | |
7VMT
 
 | |
7EA6
 
 | | Crystal structure of TCR-017 ectodomain | | Descriptor: | T cell receptor 017 alpha chain, T cell receptor 017 beta chain | | Authors: | Nagae, M, Yamasaki, S. | | Deposit date: | 2021-03-06 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.18000245 Å) | | Cite: | Identification of conserved SARS-CoV-2 spike epitopes that expand public cTfh clonotypes in mild COVID-19 patients.
J.Exp.Med., 218, 2021
|
|
8Y6X
 
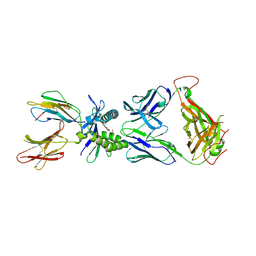 | | Crystal structure of ternary complex of human MR1, ligand #4, and MAIT-TCR A-F7 | | Descriptor: | 5-(2-oxidanylidenepropyl)-8-[(2~{S},3~{S},4~{R})-2,3,4,5-tetrakis(oxidanyl)pentyl]-1,7-dihydropteridine-2,4,6-trione, Beta-2-microglobulin, MAIT T cell receptor (A-F7) alpha chain, ... | | Authors: | Nagae, M, Inuki, S, Yamasaki, S. | | Deposit date: | 2024-02-03 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Development of Ribityllumazine Analogue as Mucosal-Associated Invariant T Cell Ligands.
J.Am.Chem.Soc., 146, 2024
|
|
5AZX
 
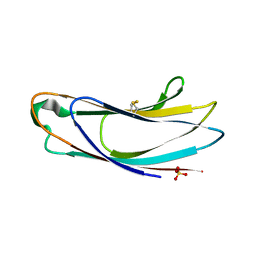 | | Crystal structure of p24delta1 GOLD domain (native 1) | | Descriptor: | SULFATE ION, Transmembrane emp24 domain-containing protein 10 | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2015-10-23 | | Release date: | 2016-09-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | 3D Structure and Interaction of p24 beta and p24 delta Golgi Dynamics Domains: Implication for p24 Complex Formation and Cargo Transport
J.Mol.Biol., 428, 2016
|
|
5AV7
 
 | | Crystal structure of Calsepa lectin in complex with bisected glycan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)][2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]methyl alpha-D-mannopyranoside, Lectin | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2015-06-12 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Atomic visualization of a flipped-back conformation of bisected glycans bound to specific lectins
Sci Rep, 6, 2016
|
|
5AZW
 
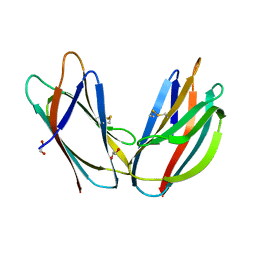 | | Crystal structure of p24beta1 GOLD domain | | Descriptor: | 1,2-ETHANEDIOL, Transmembrane emp24 domain-containing protein 2 | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2015-10-23 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 3D Structure and Interaction of p24 beta and p24 delta Golgi Dynamics Domains: Implication for p24 Complex Formation and Cargo Transport
J.Mol.Biol., 428, 2016
|
|
5AVA
 
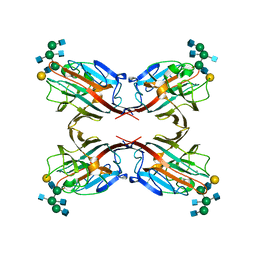 | |
5B1X
 
 | |
5B1W
 
 | |
5AZY
 
 | | Crystal structure of p24delta1 GOLD domain (Native 2) | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Transmembrane emp24 domain-containing protein 10 | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2015-10-23 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 3D Structure and Interaction of p24 beta and p24 delta Golgi Dynamics Domains: Implication for p24 Complex Formation and Cargo Transport
J.Mol.Biol., 428, 2016
|
|
2ZOT
 
 | |
2ZOU
 
 | | Crystal structure of human F-spondin reeler domain (fragment 2) | | Descriptor: | 1,2-ETHANEDIOL, Spondin-1 | | Authors: | Nagae, M, Nogi, T, Takagi, J. | | Deposit date: | 2008-06-07 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the F-spondin reeler domain reveals a unique beta-sandwich fold with a deformable disulfide-bonded loop
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2ZHM
 
 | | Crystal structure of human galectin-9 N-terminal CRD in complex with N-acetyllactosamine trimer (crystal 1) | | Descriptor: | (R)-1-PARA-NITRO-PHENYL-2-AZIDO-ETHANOL, Galectin-9, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nagae, M, Nishi, N, Murata, T, Usui, T, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain.
Glycobiology, 19, 2009
|
|
2ZHN
 
 | | Crystal structure of human galectin-9 N-terminal CRD in complex with N-acetyllactosamine trimer (crystal 2) | | Descriptor: | (R)-1-PARA-NITRO-PHENYL-2-AZIDO-ETHANOL, GALECTIN-9, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Nagae, M, Nishi, N, Murata, T, Usui, T, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain.
Glycobiology, 19, 2009
|
|
