1V87
 
 | | Solution Structure of the Ring-H2 Finger Domain of Mouse Deltex Protein 2 | | Descriptor: | Deltex protein 2, ZINC ION | | Authors: | Miyamoto, K, Muto, Y, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-12-29 | | Release date: | 2004-06-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Ring-H2 Finger Domain of Mouse Deltex Protein 2
To be Published
|
|
1A1P
 
 | | COMPSTATIN, NMR, 21 STRUCTURES | | Descriptor: | COMPSTATIN | | Authors: | Morikis, D, Assa-Munt, N, Sahu, A, Lambris, J.D. | | Deposit date: | 1997-12-12 | | Release date: | 1998-04-08 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Compstatin, a potent complement inhibitor.
Protein Sci., 7, 1998
|
|
1V5Q
 
 | | Solution Structure of the PDZ Domain from Mouse Glutamate Receptor Interacting Protein 1A-L (GRIP1) Homolog | | Descriptor: | Glutamate Receptor Interacting Protein 1A-L Homolog | | Authors: | Sato, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the PDZ Domain from Mouse Glutamate Receptor Interacting Protein 1A-L (GRIP1) Homolog
To be Published
|
|
1V35
 
 | | Crystal Structure of Eoyl-ACP Reductase with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, enoyl-ACP reductase | | Authors: | SwarnaMukhi, P.L, Kapoor, M, surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2003-10-28 | | Release date: | 2004-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the variation in triclosan affinity to enoyl reductases.
J.Mol.Biol., 343, 2004
|
|
1A0A
 
 | | PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*TP*CP*CP*CP*AP*CP*GP*TP*GP*TP*GP*AP*G )-3'), DNA (5'-D(*CP*TP*CP*AP*CP*AP*CP*GP*TP*GP*GP*GP*AP*CP*TP*AP*G )-3'), PROTEIN (PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4) | | Authors: | Shimizu, T, Toumoto, A, Ihara, K, Shimizu, M, Kyogoku, Y, Ogawa, N, Oshima, Y, Hakoshima, T. | | Deposit date: | 1997-11-27 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition.
EMBO J., 16, 1997
|
|
1A4V
 
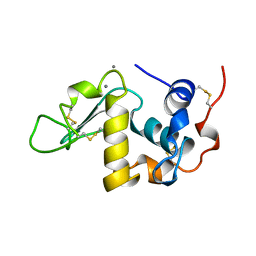 | | ALPHA-LACTALBUMIN | | Descriptor: | ALPHA-LACTALBUMIN, CALCIUM ION | | Authors: | Chandra, N, Acharya, K.R. | | Deposit date: | 1998-02-05 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evidence for the presence of a secondary calcium binding site in human alpha-lactalbumin.
Biochemistry, 37, 1998
|
|
1B2V
 
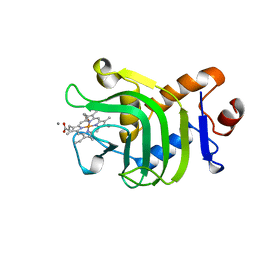 | | HEME-BINDING PROTEIN A | | Descriptor: | CALCIUM ION, PROTEIN (HEME-BINDING PROTEIN A), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnoux, P, Haser, R, Izadi, N, Lecroisey, A, Wandersma, N.C, Czjzek, M. | | Deposit date: | 1998-12-01 | | Release date: | 1999-06-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of HasA, a hemophore secreted by Serratia marcescens.
Nat.Struct.Biol., 6, 1999
|
|
1B70
 
 | | PHENYLALANYL TRNA SYNTHETASE COMPLEXED WITH PHENYLALANINE | | Descriptor: | MAGNESIUM ION, PHENYLALANINE, PHENYLALANYL-TRNA SYNTHETASE | | Authors: | Reshetnikova, L, Moor, N, Lavrik, O, Vassylyev, D.G. | | Deposit date: | 1999-01-26 | | Release date: | 2000-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of phenylalanyl-tRNA synthetase complexed with phenylalanine and a phenylalanyl-adenylate analogue
J.Mol.Biol., 287, 1999
|
|
1B3U
 
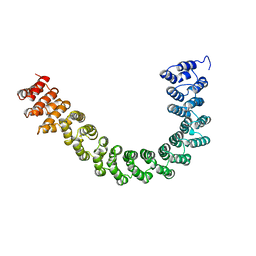 | | CRYSTAL STRUCTURE OF CONSTANT REGULATORY DOMAIN OF HUMAN PP2A, PR65ALPHA | | Descriptor: | PROTEIN (PROTEIN PHOSPHATASE PP2A) | | Authors: | Groves, M.R, Hanlon, N, Turowski, P, Hemmings, B, Barford, D. | | Deposit date: | 1998-12-14 | | Release date: | 1999-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs.
Cell(Cambridge,Mass.), 96, 1999
|
|
1BFW
 
 | | RETRO-INVERSO ANALOGUE OF THE G-H LOOP OF VP1 IN FOOT-AND-MOUTH-DISEASE (FMD) VIRUS, NMR, 10 STRUCTURES | | Descriptor: | VP1 PROTEIN | | Authors: | Petit, M.C, Benkirane, N, Guichard, G, Phan Chan Du, A, Cung, M.T, Briand, J.P, Muller, S. | | Deposit date: | 1998-05-22 | | Release date: | 1999-01-13 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a retro-inverso peptide analogue mimicking the foot-and-mouth disease virus major antigenic site. Structural basis for its antigenic cross-reactivity with the parent peptide.
J.Biol.Chem., 274, 1999
|
|
1AQP
 
 | | RIBONUCLEASE A COPPER COMPLEX | | Descriptor: | COPPER (II) ION, RIBONUCLEASE A | | Authors: | Ramasubbu, N. | | Deposit date: | 1997-07-31 | | Release date: | 1998-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the copper and nickel complexes of RNase A: metal-induced interprotein interactions and identification of a novel copper binding motif.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1V31
 
 | | Solution structure of the SWIB/MDM2 domain of the hypothetical protein At5g14170 from Arabidopsis thaliana | | Descriptor: | hypothetical protein RAFL11-05-P19 | | Authors: | Yoneyama, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-21 | | Release date: | 2004-04-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SWIB/MDM2 domain of the hypothetical protein At5g14170 from Arabidopsis thaliana
To be Published
|
|
1V9V
 
 | | Solution structure of putative domain of human KIAA0561 protein | | Descriptor: | KIAA0561 protein | | Authors: | Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-03 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of putative domain of human KIAA0561 protein
To be Published
|
|
1A05
 
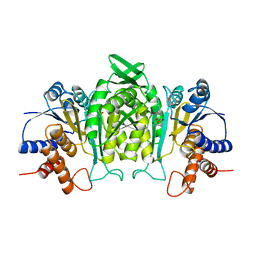 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 3-ISOPROPYLMALATE DEHYDROGENASE FROM THIOBACILLUS FERROOXIDANS WITH 3-ISOPROPYLMALATE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, 3-ISOPROPYLMALIC ACID, MAGNESIUM ION | | Authors: | Imada, K, Inagaki, K, Matsunami, H, Kawaguchi, H, Tanaka, H, Tanaka, N, Namba, K. | | Deposit date: | 1997-12-09 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of 3-isopropylmalate dehydrogenase in complex with 3-isopropylmalate at 2.0 A resolution: the role of Glu88 in the unique substrate-recognition mechanism.
Structure, 6, 1998
|
|
1AN7
 
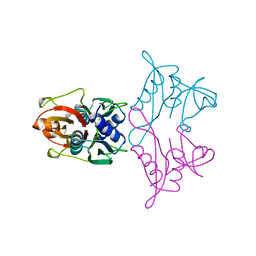 | |
1VAE
 
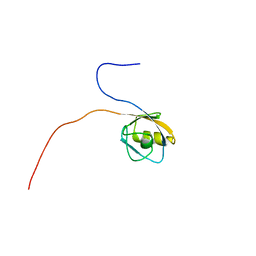 | | Solution structure of the PDZ domain of mouse Rhophilin-2 | | Descriptor: | rhophilin, Rho GTPase binding protein 2 | | Authors: | Yoneyama, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-14 | | Release date: | 2004-08-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PDZ domain of mouse Rhophilin-2
To be Published
|
|
1VBU
 
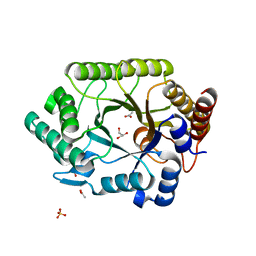 | | Crystal structure of native xylanase 10B from Thermotoga maritima | | Descriptor: | ACETIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Ihsanawati, Kumasaka, T, Kaneko, T, Nakamura, S, Tanaka, N. | | Deposit date: | 2004-03-02 | | Release date: | 2005-06-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the substrate subsite and the highly thermal stability of xylanase 10B from Thermotoga maritima MSB8
Proteins, 61, 2005
|
|
1A41
 
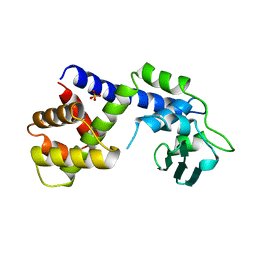 | | TYPE 1-TOPOISOMERASE CATALYTIC FRAGMENT FROM VACCINIA VIRUS | | Descriptor: | SULFATE ION, TOPOISOMERASE I | | Authors: | Cheng, C, Kussie, P, Pavletich, N, Shuman, S. | | Deposit date: | 1998-02-10 | | Release date: | 1999-06-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases.
Cell(Cambridge,Mass.), 92, 1998
|
|
3AI4
 
 | | Crystal structure of yeast enhanced green fluorescent protein - mouse polymerase iota ubiquitin binding motif fusion protein | | Descriptor: | SULFATE ION, yeast enhanced green fluorescent protein,DNA polymerase iota | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallization of small proteins assisted by green fluorescent protein
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1B0Q
 
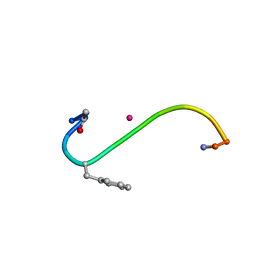 | | DITHIOL ALPHA MELANOTROPIN PEPTIDE CYCLIZED VIA RHENIUM METAL COORDINATION | | Descriptor: | PROTEIN (CYCLIC ALPHA MELANOCYTE STIMULATING HORMONE), RHENIUM | | Authors: | Giblin, M.F, Wang, N, Hoffman, T.J, Jurisson, S.S, Quinn, T.P. | | Deposit date: | 1998-11-12 | | Release date: | 1998-11-18 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of alpha-melanotropin peptide analogs cyclized through rhenium and technetium metal coordination.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
2ZZJ
 
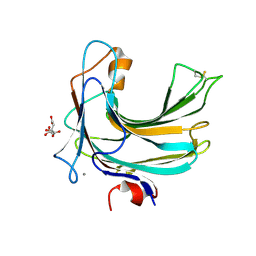 | | Crystal structure of endo-beta-1,4-glucuronan lyase from fungus Trichoderma reesei | | Descriptor: | CALCIUM ION, CITRIC ACID, Glucuronan lyase A | | Authors: | Konno, N, Ishida, T, Fushinobu, S, Igarashi, K, Habu, N, Samejima, M, Isogai, A. | | Deposit date: | 2009-02-16 | | Release date: | 2009-05-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of polysaccharide lyase family 20 endo-beta-1,4-glucuronan lyase from the filamentous fungus Trichoderma reesei.
Febs Lett., 583, 2009
|
|
1V5S
 
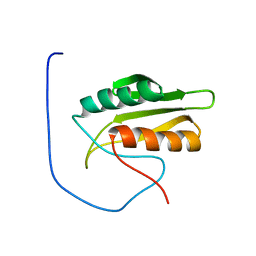 | | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3 | | Descriptor: | MAP/microtubule affinity-regulating kinase 3 | | Authors: | Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3
To be Published
|
|
1V5H
 
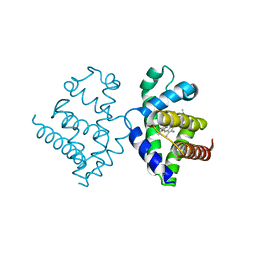 | | Crystal Structure of Human Cytoglobin (Ferric Form) | | Descriptor: | Cytoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugimoto, H, Makino, M, Sawai, H, Kawada, N, Yoshizato, K, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of human cytoglobin for ligand binding.
J.Mol.Biol., 339, 2004
|
|
1V95
 
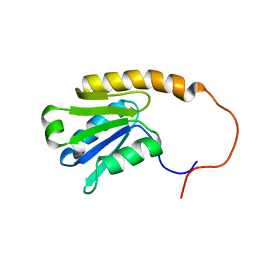 | | Solution Structure of Anticodon Binding Domain from Nuclear Receptor Coactivator 5 (Human KIAA1637 Protein) | | Descriptor: | Nuclear receptor coactivator 5 | | Authors: | Zhao, C, Kigawa, T, Tochio, N, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-20 | | Release date: | 2004-07-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Anticodon Binding Domain from Nuclear Receptor Coactivator 5 (Human KIAA1637 Protein)
To be Published
|
|
3AI5
 
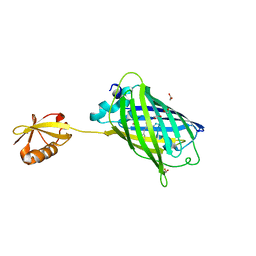 | | Crystal structure of yeast enhanced green fluorescent protein-ubiquitin fusion protein | | Descriptor: | 1,2-ETHANEDIOL, yeast enhanced green fluorescent protein,Ubiquitin | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallization of small proteins assisted by green fluorescent protein
Acta Crystallogr.,Sect.D, 66, 2010
|
|
