6WR3
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J36 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*GP*AP*CP*AP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-04-29 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.173 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSN
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J7 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*GP*AP*CP*GP*TP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WRC
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J31 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*GP*AP*CP*GP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), DNA (5'-D(P*CP*GP*TP*CP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-04-29 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.178 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSU
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J19 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*AP*GP*AP*CP*TP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSQ
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J10 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*TP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSY
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J23 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*AP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.053 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSV
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J20 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*GP*AP*CP*AP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSS
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J15 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*GP*AP*CP*CP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
5UDN
 
 | |
6WT0
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J25 immobile Holliday junction | | Descriptor: | COBALT (II) ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*AP*GP*AP*CP*CP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSP
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J9 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*GP*AP*CP*TP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.049 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WT1
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J29 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*AP*AP*GP*AP*CP*TP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), DNA (5'-D(P*AP*GP*TP*CP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
8SF9
 
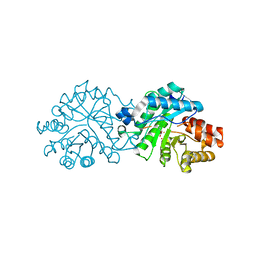 | | Crystal structure of the engineered SsoPox variant IG7 - Alternative state | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
6WSR
 
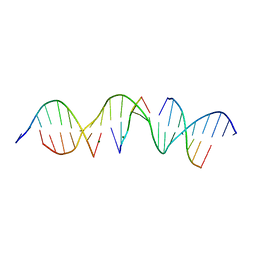 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J10 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*AP*GP*AP*CP*CP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), DNA (5'-D(P*CP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSZ
 
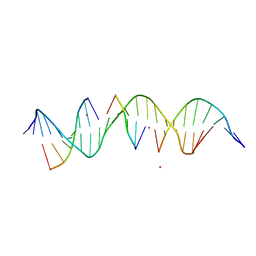 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J24 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*GP*AP*CP*TP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.053 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSW
 
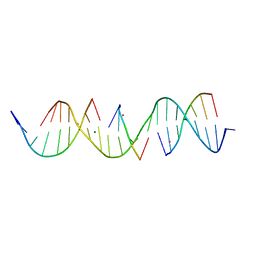 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J21 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*AP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*TP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSX
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J22 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*GP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), DNA (5'-D(P*CP*GP*TP*CP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.122 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
8SFA
 
 | | Crystal structure of the engineered SsoPox variant IIIC1 | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SFW
 
 | | Crystal structure of TuUGT202A2 (Tetur22g00270) in complex with quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, UDP-glycosyltransferase 202A2, URIDINE-5'-DIPHOSPHATE | | Authors: | Arriaza, R.H, Dermauw, W, Wybouw, N, Van Leeuwen, T, Chruszcz, M. | | Deposit date: | 2023-04-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of TuUGT202A2 (Tetur22g00270) in complex with quercetin
To Be Published
|
|
6F4G
 
 | | 'Crystal structure of the Drosophila melanogaster SNF/U2A'/U2-SL4 complex | | Descriptor: | CHLORIDE ION, Probable U2 small nuclear ribonucleoprotein A', SULFATE ION, ... | | Authors: | Weber, G, DeKoster, G, Holton, N, Hall, K.B, Wahl, M.C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular principles underlying dual RNA specificity in the Drosophila SNF protein.
Nat Commun, 9, 2018
|
|
8SFC
 
 | | Crystal structure of the engineered SsoPox variant IVA4 in alternate state | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SFK
 
 | | Crystal structure of the engineered SsoPox variant IVE2 | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
5UBF
 
 | |
8SFD
 
 | | Crystal structure of the engineered SsoPox variant IVB10 | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
6F2T
 
 | | Crystal structure of ectonucleotide phosphodiesterase/pyrophosphatase-3 (NPP3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dohler, C, Zebisch, M, Strater, N. | | Deposit date: | 2017-11-27 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and substrate binding mode of ectonucleotide phosphodiesterase/pyrophosphatase-3 (NPP3).
Sci Rep, 8, 2018
|
|
